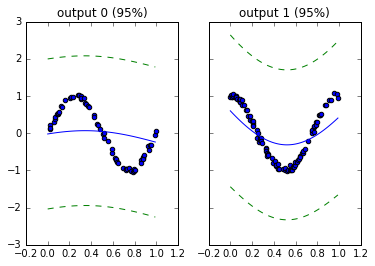
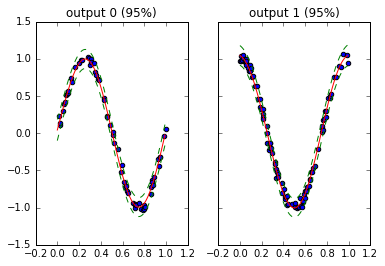
Do you like to apply Bayesian nonparameteric methods to your regressions? Are you frequently tempted by the flexibility that kernel-based learning provides? Do you have trouble getting structured kernel interpolation or various training conditional inducing point approaches to work in a non-stationary multi-output setting?
If so, this package is for you.
runlmc is a Python 3.5+ package designed to extend structural efficiencies from Scalable inference for structured Gaussian process models (Staaçi 2012) and Thoughts on Massively Scalable Gaussian Processes (Wilson et al 2015) to the non-stationary setting of linearly coregionalized multiple-output regressions. For the single output setting, MATLAB implementations are available here.
In other words, this provides a matrix-free implementation of multi-output GPs for certain covariances. As far as I know, this is also the only matrix-free implementation for single-output GPs in python.
- Zero-mean only for now.
- Check out the latest documentation
- Check out the Dev Stuff section below for installation requirements.
- Accepts arbitrary input dimensions are allowed, but the number of active dimensions in each kernel must still be capped at two (though a model can have multiple different kernels depending on different subsets of the dimensions).
GPy is a way more general GP library that was a strong influence in the development of this one. I've tried to stay as faithful as possible to its structure.
I've re-used a lot of the GPy code. The main issue with simply adding my methods to GPy is that the API used to interact between GPy's kern, likelihood, and inference packages centers around the dL_dK object, a matrix derivative of the likelihood with respect to covariance. The materialization of this matrix is the very thing my algorithm tries to avoid for performance.
If there is some quantifiable success with this approach then integration with GPy would be a reasonable next-step.
n_per_output = [65, 100]
xss = list(map(np.random.rand, n_per_output))
yss = [f(2 * np.pi * xs) + np.random.randn(len(xs)) * 0.05
for f, xs in zip([np.sin, np.cos], xss)]
ks = [RBF(name='rbf{}'.format(i)) for i in range(nout)]
ranks = [1]
fk = FunctionalKernel(D=len(xss), lmc_kernels=ks, lmc_ranks=ranks)
lmc = LMC(xss, yss, functional_kernel=fk)
# ... plotting code
lmc.optimize()
# ... more plotting code
For runnable code, check examples/.
Make sure that the directory root is in the PYTHONPATH when running the benchmarks. E.g., from the directory root:
PYTHONPATH=.. jupyter notebook examples/example.ipynb
cd benchmarks/fx2007 && ./run.sh # will take a while!
All below invocations should be done from the repo root.
| Command | Purpose |
|---|---|
./style.sh |
Check style with pylint, ignoring TODOs and locally-disabled warnings. |
./docbuild.sh |
Regenerate docs (index will be in doc/_generated/_build/index.html) |
nosetests |
Run unit tests |
./arxiv-tar.sh |
Create an arxiv-friendly tarball of the paper sources |
python setup.py install |
Install minimal runtime requirements for runlmc |
./asvrun.sh |
run performance benchmarks |
To develop, requirements also include:
sphinx sphinx_rtd_theme matplotlib codecov pylint parameterized pandas contexttimer GPy asv
To build the paper, the packages epstool and epstopdf are required. Developers should also have sphinx sphinx_rtd_theme matplotlib GPy codecov pylint parameterized pandas contexttimer installed.
- Make
standard_testerstale-tolerable: can't fetch data, code fromgithubwithout version inconsistency. - Make
grad-gridbenchmark only generatepdffiles directly, get rid ofepstool,epstopdfdeps. - Make all benchmarks accept --validate (And add --validate test for representation-cmp : inv path should be tested in
bench.py) - Automatically trigger ./asvrun.sh on commit, somehow
- Automatically find
min_grad_ratioparameter / get rid of it.- Logdet approximations: (1) Chebyshev-Hutchinson Code (2) Integral Probing (3) Lanczos (4) approx
tr log (A)with MVM from f(A)b paper
- Logdet approximations: (1) Chebyshev-Hutchinson Code (2) Integral Probing (3) Lanczos (4) approx
- Preconditioning
- Cache Krylov solutions over iterations?
- Cutajar 2016 iterative inversion approach?
- T.Chan preconditioning for specialized on-grid case (needs development of partial grid)
- TODO(test) - document everything that's missing documentation along the way.
- Current prediction generates the full covariance matrix, then throws everything but the diagonal away. Can we do better?
- Compare to MTGP, CGP
- Minor perf improvements: what helps?
- CPython; numba.
- In-place multiplication where possible
- square matrix optimizations
- TODO(sparse-derivatives)
- bicubic interpolation: invert order of xs/ys for locality gains (i.e., interpolate x first then y)
- TODO(sum-fast) low-rank dense multiplications give SumKernel speedups?
- multidimensional inputs and ARD.
- TODO(prior). Compare to spike and slab, also try MedGP (e.g., three-parameter beta) - add tests for priored versions of classes, some tests in parameterization/ (priors should be value-cached, try to use an external package)
- HalfLaplace should be a Prior, add vectorized priors (remembering the shape)
- Migrate to asv, separate tests/ folder (then no autodoc hack to skip test_* modules; pure-python benchmarks enable validation of weather/ and fx2007 benchmarks on travis-ci but then need to be decoupled from MATLAB implementations)
- mean functions
- product kernels (multiple factors)
- active dimension optimization
- Consider other approximate inverse algorithms: see Thm 2.4 of Agarwal, Allen-Zhu, Bullins, Hazan, Ma 2016