-
Notifications
You must be signed in to change notification settings - Fork 6
Development subsites
Subsite (lab) landing pages are built and served from the Galaxy Media Site under
/landing/<lab-name>. GMS has a framework for building new labs pages in a
consistent and reproducible manner, with a schema written in Python.
- Create a git branch for the new lab.
- Decide on a consisteny lab name to use. This will be referred to as
$LABhere. - Create a new subsite dir by cloning
webapp/home/subdomains/genome/towebapp/home/subdomains/${LAB}. - Create webpage content for the new lab by modifying the
${LAB}/sections.py${LAB}/content/*.pyfiles, observing the Python schema (see below). - Clone the HTML template
webapp/home/templates/home/subdomains/genome.htmlto${LAB}.html. - Edit the HTML template to make the landing page for your lab. It will render the content created in the step above as sections/accordion elements.
- You may wish to reuse/create snippets for this template in
webapp/home/templates/home/subdomains/components. - Run the branch with local development server.
- In the web admin, add a
Subsiterecord with the lab name$LAB. - You should find the lab landing page at the URL
/landing/$LAB.
Content for the lab pages is defined in a Python schema (ideally this would be YAML, but since YAML can't handle variables natively this would lead to large/repetitive files.)
A basic understanding of Python is required to contribute to this content, but can be done by a non-developer (especially if they have Git familiarity). Ideally this will be the domain expert responsible for designing the page.
The Python schema goes something like this:
# As defined in sections.py:
sections = [
{
"id": str,
"title": str, # e.g. "Data import and preparation"
"tabs": [
{
"id": str, # e.g. "tools"
"title": str, # e.g. "Tools"
"heading_md": str, # accepts inline HTML e.g. <code> <b> <i> <a>
"content": content,
},
# ... Each of the above will make a new tab in this section
],
},
# ... Each of the above will make a new section
]
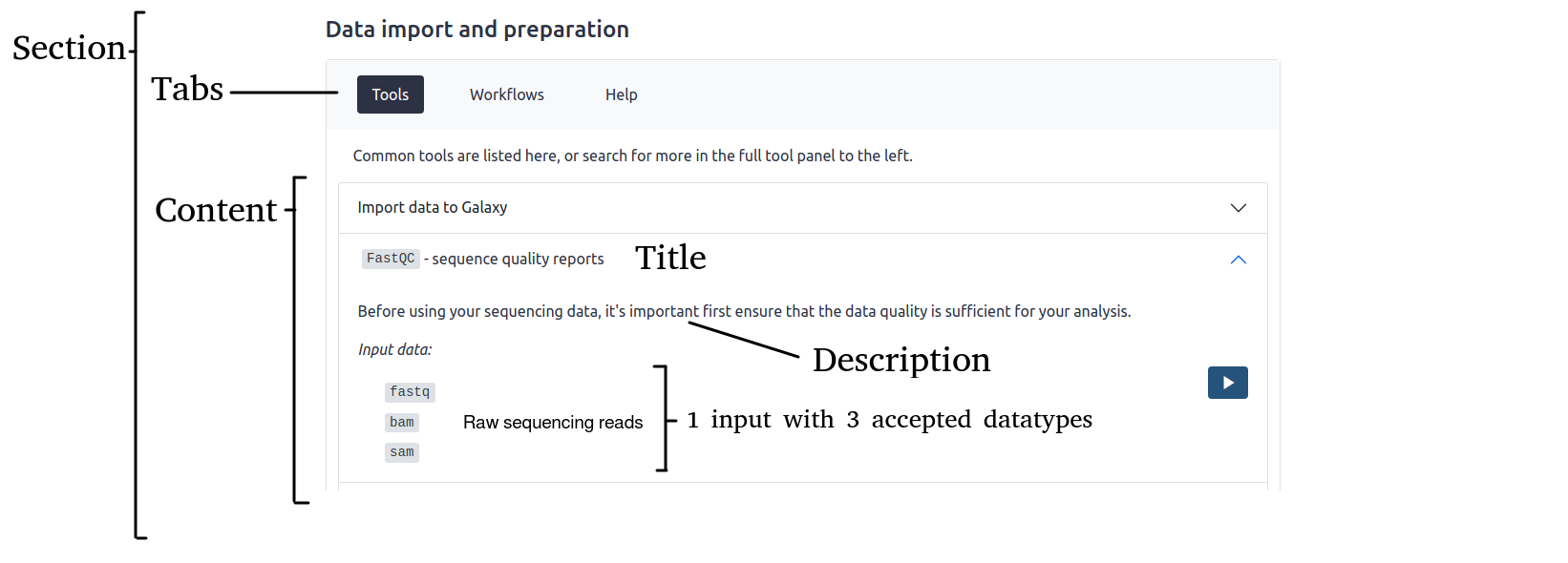
# As defined in content/*.py:
content = [
{
"title_md": str, # inline HTML
"description_md": str, # inline HTML; should be a <p>; can have internal line breaks etc.
"inputs": [
{
'datatypes': [str, ], # List of accepted Galaxy datatypes for this input
'label': str, # Name of this input
},
{ # An example input:
'datatypes': ['fastq', 'bam', 'sam'], # List of accepted datatypes for this input
'label': 'Raw sequencing reads', # Descriptive name for this input
},
# ...
],
# Optional fields:
"button_link": str, # URL for run button e.g. Galaxy tool link
"button_md": str, # defaults to "play" icon
"button_tip": str, # tooltip text; defaults to "Run tool"
"view_link": str, # URL for view button e.g. workflowhub link
"view_md": str, # defaults to "eye" icon
"view_tip": str, # tooltip text; defaults to None
},
# ... Each of the above will make an accordion item
]Some sections (e.g. "Help") may benefit from having a longer description with more inline HTML, for example:
{
"title_md": 'Can I use Apollo to share and edit the annotated genome?',
"description_md": """
<p>
Apollo is web-browser accessible system that lets you conduct real-time collaborative curation and editing of genome annotations.
</p>
<p>
The Australian BioCommons and our partners at QCIF and Pawsey provide a hosted
<a href="https://apollo-portal.genome.edu.au/" target="_blank">
Apollo Portal service
</a>
where your genome assembly and supporting evidence files can be hosted. All system administration is taken care of, so you and your team can focus on the annotation curation itself.
</p>
<p>
This
<a href="https://training.galaxyproject.org/training-material/topics/genome-annotation/tutorials/apollo-euk/tutorial.html" target="_blank">
Galaxy tutorial
</a>
provides a complete walkthrough of the process of refining eukaryotic genome annotations with Apollo.
</p>
""",
"button_link": "https://support.biocommons.org.au/support/solutions/articles/6000244843-apollo-for-collaborative-curation-and-editing",
"button_md": "More info",
}The button_link or view_link can be omitted to hide that button.
The above schema will render a webpage that looks something like this:
For full examples of the above, please refer to genome/content/assembly.py.