Providing insight into population displacement during natural disasters
- Set up conda environment
$ conda env create -f environment.yml
$ conda activate COvid19
- Set up automatic downloader
- Download latest Google Chrome driver to
automate_download/ - Edit
automate_download/credential.json, using your facebook email and password
- Find target of interest on Facebook GeoInsights Portal
- Click Facebook Population (Tile Level) and then
Download. Save the url of thisDownloadpage. - Edit
automate_download/config.json- Ensure correctness of
downloads - Enter url of facebook webpage for Tile Level downloads into
tile_link - Edit
repoto the correct path
/pop_tiles/{target_name}_pop_tiles/orig/ - Ensure correctness of
- Run
$ python scrape.py --config config.json --cred --credential.json
- Note: Often the this script will place the downloaded files in your
downloadsfolder. If so, manually move the files to therepodefined in yourconfig.json
- Edit
snake_config.json- Using the defined format, create a new entry. (It is recommended you copy and paste a previously defined city as the formatting is crucial)
- The json key,
sit_rep_name,county_name, andcity_nameshould all be the same name. This name should be one word, begin with a capitol letter and define the target of interest dbshould establish the path to your database (not yet built), this database should be the same as yoursit_rep_nameexcept all lower casecitiesis always_(later functionality)- Download the
county_shapes - The pipeline requires shape file data of the region of interest.
- The easiest way to obtain this data is to google
{region} + shape file. (Example: 'Lebanon shape file') - Yields : https://data.humdata.org/dataset/lebanon-administrative-boundaries-levels-0-3
- In this example we can see the shape file data for administrative boundaries level 0, 1 ,2 and 3 for Lebanon
- Upon downloading this we see that it includes many files, we want level 0 in the following file formats
.shx,.shp.xml,.shp,.sbx,.sbn,.prj,.dbf,.cpg- Select these files for level 0 and move them into a new directory in
facebook/shapefiles/... - Open a new terminal and
cd facebook/shapefiles/ - Run
$ python >>> import geopandas as gpd >>> gdf = gpd.read_file("{shape_directory}") >>> gdf - This should output a table containing a
geometrycolumn and another column that will contain either cities, districts, villages, counties, states, etc. - This column name will be your
county_shapes_name city_shapesis always_(later functionality)city_shapes_nameis always_(later functionality)- If you are researching a very large dataset it is highly recommended that you identify a sub-region of interest. If you choose to do such simply draw a coordinate box
and input the four respective coordinates into
min_lat,max_lat,min_lon, andmax_lonrespectively. - If you would like to research the entire dataset then input
NONEinto all four categories. - Finally input the population tiles into
Repo.
- Edit
init.json - This config file will establish the targets of interest you would like to investigate concurenntly.
- You may add as many as targets as you wish (Note: Only four concurrent cities have been tested at this point). Be mindful of the collective data useage your targets will demand.
- Enter the
sit_rep_nameascity - Enter the start date of interest as
start_date - Enter the end date of interest as
end_date - It is very important you follow the defined format of
YYYY,M,D. Do no prepend single digit numbers with a zero. - Define your system path to the pipeline in
path
Setup is now complete!
- Ensure the conda environment is activated
- Run
$ python __init__.py
- If successful you will have a directory for each of you defined targets in
/dash/saved_data/- In each of these new directories you will have five files
-
animation.pickle,dates_times.pickle,json_geo.json,pre_graph_data.csv, andtrend_lines.pickle
- In each of these new directories you will have five files
-
- From
/dashRun
$ python local_dash.py
- Go to
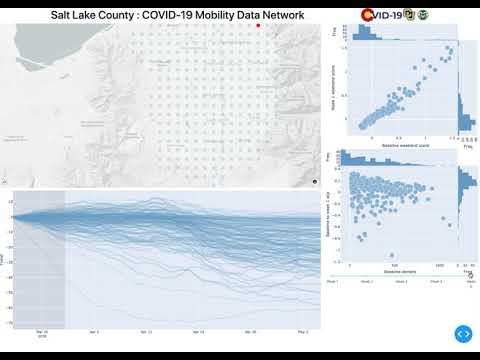
http://127.0.0.1:8050/ - The initial page is the index page
- These graphs display each of the requested targets of interest, with the corresponding number of tiles that deviated away from the median of that day.
- Using this information the researcher can decide where anomalities warrant further research, in which case you can click the hyperlink above each graph
- You may also change the url directly with your
sit_rep_name(http://127.0.0.1:8050/Lebanon)
- You may also change the url directly with your
- This will redirect you to an indepth profile on your target of interest
- The map contains an animation frame for each day
- The line plots are the trend component of the season decompositon
- Each line is attached to a tile. You can use the plotly selection tools to isolate tiles and lines of further interest
$ python src/csv_to_sql.py --csv pop_tiles/ --db colorado.db
rm -f colorado.db;sqlite3 colorado.db < tmp.sql
$ rm -f colorado.db;sqlite3 colorado.db < tmp.sql
- Get scores aggregated by a field in a shapefile, here we will use cities in Colorado.
$ python src/get_all_scores_by_shape.py \
--db colorado.db \
--shapefile shapefiles/Colorado_City_Boundaries/ \
--shapename NAME10 \
> colorado_city_scores.`date "+%Y%m%d"`.txt
On March 24th, 2020 at 1800 MNT, our team started collecting data from Facebook Data for Good (https://dataforgood.fb.com/) that covers Utah and Colorado. Facebook provides aggregated and anonymized user density data for 2km x 2km tiles at three timepoints per day (0200 MNT, 1000 MNT, and 16000 MNT). To help protect privacy, density data is not reporting when a tile has fewer than 10 users. In addition to the current density data, which is referred to as the crisis density, Facebook also gives the baseline density for each tile and time point that is the average density of that tile at the day and time for the 6 weeks prior to the start of the crisis (March 25th, 2020).
With 78 observations per tile and counting, we can track changes in population density over time to infer behavior. For example, comparing crisis and baseline densities can show how patterns have changed after stay-at-home orders were issued and comparing one crisis week to the next can show if the new patterns are holding.
 Baseline (blue) and current (orange) population densities for one tile in Salt
Lake City. The current data starts on Tuesday, March 24, 2020, at 1800 MNT. The
last time point shown is March 18, 2020, at 1000 MNT, but we continue to
collect new data. Each day (shaded grey) includes time points at 0200, 1000,
and 1800. Weekends are darkened.
Baseline (blue) and current (orange) population densities for one tile in Salt
Lake City. The current data starts on Tuesday, March 24, 2020, at 1800 MNT. The
last time point shown is March 18, 2020, at 1000 MNT, but we continue to
collect new data. Each day (shaded grey) includes time points at 0200, 1000,
and 1800. Weekends are darkened.
python src/plot_one.py \
-i colorado_city_scores.20200422.txt \
-o imgs/Boulder_example_tile.png \
--shapename "Boulder" \
--width 5 -n 5 --height 1
Most regions show a regular weekly pattern. Economic centers are denser on the
weekdays, residential areas are denser on the weekends, and recreational areas
are more consistent. To measure these differences we developed the weekend
score that
compares the average weekday density
to the average weekend density
Areas with higher positive values have more weekday activity and areas with lower negative values have more weekend activity.
 Baseline and current population densities for an economic center (top, baseline ws = 1.48 ), a residential area (middle, baseline ws = -0.21), and a ski area (bottom, baseline ws = -0.06). Average weekday and weekend activity levels are given by horizontal black lines.
Baseline and current population densities for an economic center (top, baseline ws = 1.48 ), a residential area (middle, baseline ws = -0.21), and a ski area (bottom, baseline ws = -0.06). Average weekday and weekend activity levels are given by horizontal black lines.
$ python src/ws_scores.py \
-i colorado_city_scores.20200422.txt \
| sort -t$'\t' -k 5,5g \
| tail -n 1
449 Boulder 40.069664004427 -105.21606445312 1.0568428725333567 0.5266777645208203 0.5424389724825188 0.4402530723582583
$ python src/ws_scores.py \
-i colorado_city_scores.20200422.txt \
| sort -t$'\t' -k 5,5gr \
| tail -n 1
1322 Grand Lake 40.254376084285 -105.83129882812 -0.5717760132754274 -0.12254129829560738 0.005638648006131584 -0.011023686029626615
$ python src/ws_scores.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_example_tile_ws_diffs.png \
-n 449,1322 \
--width 5 --height 2.5 > /dev/null
We can track changes in behavior by comparing the scores between weeks.
 Weekend score distributions for baseline, week 1, week 2, and week 3.
Weekend score distributions for baseline, week 1, week 2, and week 3.
$ (python src/ws_scores.py -i colorado_city_scores.20200422.txt | cut -f5 | paste -sd '\t' - ; \
python src/ws_scores.py -i colorado_city_scores.20200422.txt | cut -f6 | paste -sd '\t' - ; \
python src/ws_scores.py -i colorado_city_scores.20200422.txt | cut -f7 | paste -sd '\t' - ; \
python src/ws_scores.py -i colorado_city_scores.20200422.txt | cut -f8 | paste -sd '\t' - ;) \
| src/hist.py \
-x 'Baseline ws,Week 1 ws,Week 2 ws,Week 3 ws' \
-y 'Freq' \
-b 50 \
--width 8 \
--height 2 \
-o imgs/colorado_ws_baseline_hist.png
We can also track the change in the behavior of individual tiles. In the plots below, each point is a tile and its size corresponds to the mean baseline density. Points further to the left or higher indicate more weekday activity. The closer a point is to the red trend line, the more similar the activity between the two weeks. This plot also shows a change from the baseline toward more consistent behavior between the weekdays and weekends.
 |
 |
 |
|---|
A comparison of how weekend scores change between weeks. Here, each point is a tile and the point size corresponds to the population density of the tile. Larger points are denser than smaller points. Larger positive values (right and up) indicated more weekday activity and negative values (left and down) indicate more weekend activity. The red diagonal line is a reference to judge the change between weeks. The closer a point is to the line the more similar the weekday/weekend behavior was between the weeks.
$ python src/plot_ws_x_ws.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_ws_baseline_week1.png \
--x_axis 0 \
--y_axis 1 \
-x 'Baseline ws' \
-y 'Week 1 ws' \
--width 2 \
--height 2 \
--alpha 0.25
$ python src/plot_ws_x_ws.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_ws_week1_week2.png \
--x_axis 1 \
--y_axis 2 \
-x 'Week 1 ws' \
-y 'Week 2 ws' \
--width 2 \
--height 2 \
$ python src/plot_ws_x_ws.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_ws_week2_week3.png \
--x_axis 2 \
--y_axis 3 \
-x 'Week 2 ws' \
-y 'Week 3 ws' \
--width 2 \
--height 2 \
--alpha 0.25
The behavior of a region after a stay-at-home order is issued depends on the
region’s demographics, which complicates monitoring. For example, economic
centers will be less dense, and residential centers will be denser since most
people are not working or traveling. To track how well regions are adhering to
their new patterns we developed a slip score
.
This metric assumes that adherence to these orders is best immediately
following their issuance, then tracks changes by comparing the average weekly
density
of consecutive weeks,
Areas with higher positive values are more active in the second week, and areas with lower negative values are less active.
 Current population densities for two tiles. Average weekly activity levels given by black bars. The slip from week one to week two in the top plot is 0.26 and in the slip for the same interval in the bottom plot is -0.12.
Current population densities for two tiles. Average weekly activity levels given by black bars. The slip from week one to week two in the top plot is 0.26 and in the slip for the same interval in the bottom plot is -0.12.
$ python src/slip_scores.py -i colorado_city_scores.20200422.txt | sort -t$'\t' -k 6,6gr | tail -n 2
898 Denver 39.85072040685 -104.64477539062 -1.6940531813681654 -0.34439512740216777 -0.14734599550209554
75 Denver 39.85072040685 -104.66674804688 -1.7213356921544296 -0.36289944951227465 -0.15109396965145458
$ python src/slip_scores.py -i colorado_city_scores.20200422.txt | sort -t$'\t' -k 6,6g | tail -n 2
1433 Estes Park 40.338169932441 -105.52368164062 -0.36696575466368675 0.13152468685241248 0.020371124969622872
939 Fort Garland 37.431249993417 -105.43579101562 0.18350282216948896 0.1715863040811286 -0.014918557314476904
$ python src/slip_scores.py \
-i colorado_city_scores.20200422.txt \
-o imgs/coloreado_example_slip_scores.png \
-n 75,1433 \
--width 5 \
--height 1.5
To track individual regions, we can visualize the slip score with respect to the region size. In the plots below, each point is a tile, and points from denser regions are to the right are. Points above the zero line have slipped into a more active state, and points below the line have become less active.
 |
 |
|---|
Slip scores from week 1 to week 2 (left) and from week 2 to week 3 (right). Positive values indicate an increase in week-to-week activity and negative values indicate a decrease.
$ python src/plot_split_x_density.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_ss_week1_week2.png \
--alpha 0.25 \
--width 3 \
--height 2 \
--weeks 1,2 \
-x 'Baseline density' \
-y 'Week 1 to week 2 slip'
$ python src/plot_split_x_density.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_ss_week2_week3.png \
--alpha 0.25 \
--width 3 \
--height 2 \
--weeks 2,3 \
-x 'Baseline density' \
-y 'Week 2 to week 3 slip'
Density data can be represented as a time series with repeating short-term patterns (seasons) and the overall value increases or decreases (trends). Using a model, we can decompose a series into its seasons and trends, which provides a means to compare tiles with different weekday/weekend activities (seasons).
 Density data from four tiles decomposed into their patterns and trends.
Density data from four tiles decomposed into their patterns and trends.
$ python src/trends.py -i colorado_city_scores.20200422.txt | sort -t$'\t' -k 5,5g | tail -n 2
285 Denver 39.749433173953 -104.99633789062 88.32601055634723 88.32601055634723
74 Denver 39.85072040685 -104.66674804688 128.1883336281206 128.1883336281206
$ python src/trends.py -i colorado_city_scores.20200422.txt | sort -t$'\t' -k 5,5gr | tail -n 2
1305 Aurora 39.71563761708 -104.79858398438 -21.203709164342058 21.203709164342058
494 Colorado Springs 38.831149294861 -104.75463867188 -26.088910442348492 26.088910442348492
$ python src/trends.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_example_season_trend_split.png \
-n 285,74,1305,494 \
--width 5 --height 4 > /dev/null
Even within a city, we can observe the differences in density dynamics between the business centers, which should be less dense during a stay-at-home order, and residential centers, which should be denser. To capture the overall trend of a city, we take the mean of each tile’s trend weighted by the tile’s baseline density.
 |
|---|
- Tile trends for two cities. Blue lines trend toward less density and red lines trend toward more. The line thickness corresponds to the tile’s baseline density. The black line is the mean trend weighted by population density. *
$ python src/plot_shape_trend.py \
-i colorado_city_scores.20200422.txt \
--shapename "Denver" \
-o imgs/Denver_trend.png \
--width 5 \
--height 2 \
--title "Denver"
$ python src/plot_shape_trend.py \
-i colorado_city_scores.20200422.txt \
--shapename "Colorado Springs" \
-o imgs/Colorado_Springs_trend.png \
--width 5 \
--height 2 \
--title "Colorado Springs"
The mean trends for cities can be combined for a county-level view.
County-level trends. Each line is the weighted average for tiles trends in a city. Blue lines trend toward less density and red lines trend toward more. The line thickness corresponds to the tile’s baseline density.
$ boulder_co_cities="Boulder,Erie,Jamestown,Lafayette,Longmont,Louisville,Lyons,Nederland,Superior,Ward"
$ python src/plot_shapes_mean_trend.py \
-i colorado_city_scores.20200422.txt \
--shapenames "$boulder_co_cities" \
-o imgs/boulder_co_cities_mean_trends.png \
--width 3 \
--height 2
Assuming that stay-at-home adherence was at its best the week after an order was issued (week 1), then we define "hot spots" as regions with significantly higher density levels relative to that baseline. To reduce the effects of outliers we use a 3-day mean, and to control for weekend/weekday effects we match days of the week when comparing current and week one values.
The hot-spot ()
score for a tile is the difference between the current 3-day mean
(
)
and the 3-day mean from the same starting day in the first week
(
)
divided by standard deviation of 3-day means for other similarly dense tiles
(
).
Hot spot scores for three tiles. The hot spot score (blue) is the difference between the current 3-day mean(black horizontal) and the week one 3-day mean with the same start day divided by the variance (black vertical). The three examples here show a hot spot (a significant increase over week one), a neutral spot (no significant change from week one), and a cold spot (a significant decrease over week one).
python src/hot_spot_scores.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_hotspot_scores.png \
-n 938,720,54 \
--width 5 \
--height 3 \
--ymin -7 \
--ymax 7
We are using a broader set of tiles to calculate
because we have been collecting data for only a few weeks, and we have
relatively few observations for a given tile. To increase the number of samples
used to calculate the standard deviation, we fit a linear model to the mean
density and standard deviation of all tiles, then use that model to predict the
variance for the current tile.
The mean and standard deviation of all tiles, and the linear model fit to the data.
python src/density.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_mean_x_variance.png \
--height 2 \
--width 7
Hot spot scores can either as plotted as a histogram, with respect to baseline density, or as a map (via shapefile).
 |
 |
 |
|---|
Hot-spot scores for every tile. Positive values indicate that the current density is higher today than it was in the first week. Negative values indicate the current density is less. Left: Plotted versus baseline density. Right: Viewed as as shapefile
python src/hot_spot_scores.py \
-i colorado_city_scores.20200422.txt \
| cut -f6 \
| paste -sd '\t' - \
| src/hist.py \
-o imgs/colorado_hot_spot_hist.png \
-b 50 \
-x 'Hot spot score' \
-y 'Freq' \
--width 2 --height 2
python src/plot_hot_spot_x_density.py \
-i colorado_city_scores.20200422.txt \
-o imgs/colorado_hot_spot.png \
--alpha 0.25 \
--width 3 \
--height 2 \
-x 'Baseline density' \
-y 'Current hot-spot score'
python src/make_hot_spot_shapefile.py \
-i colorado_city_scores.20200422.txt \
-o colorado_hostpot
We have also developed an interactive data browser that links these metrics to
maps using plotyly dash. Code and details on
getting the server running are in the dash folder.
Here is a short demo video