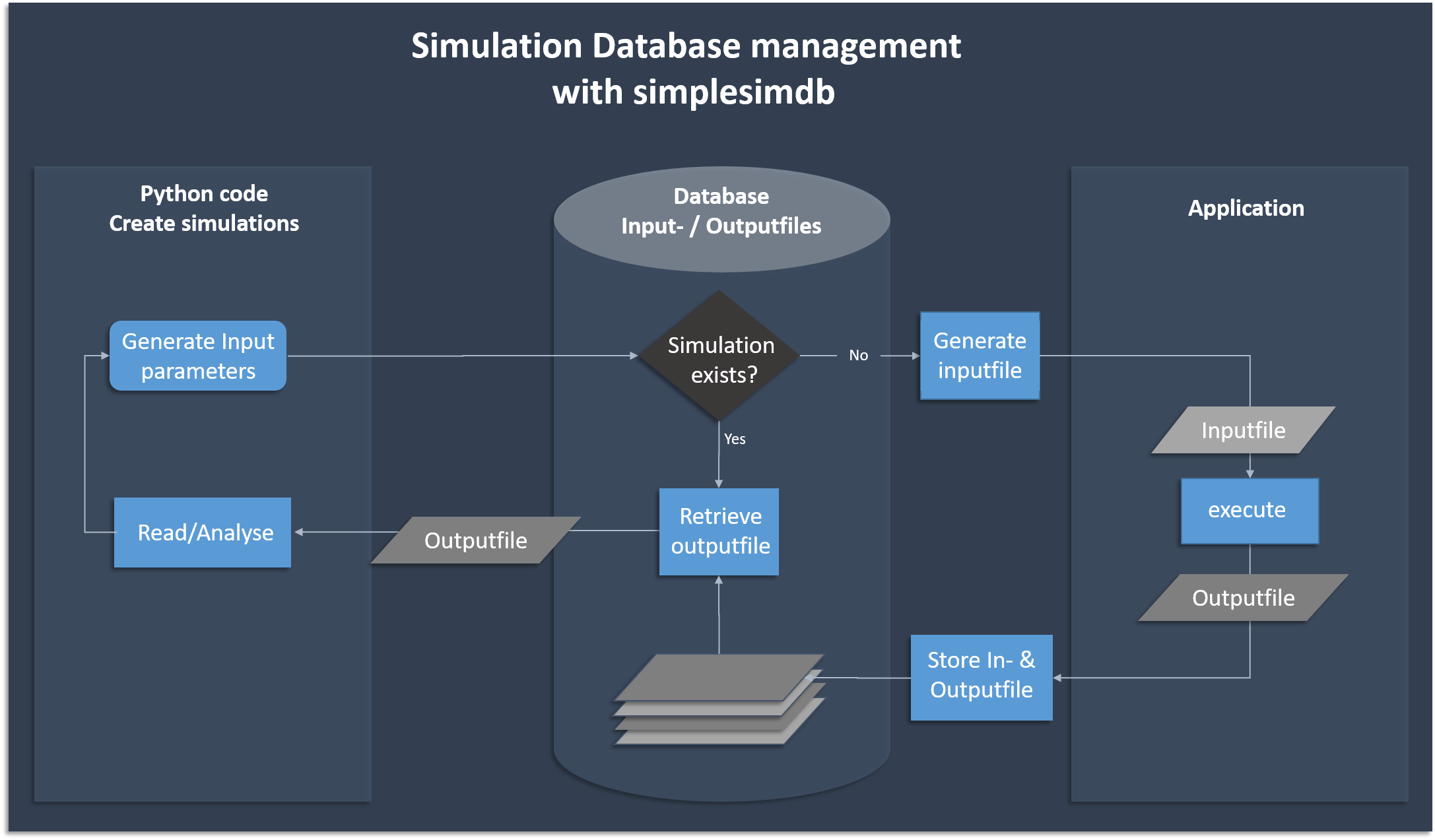
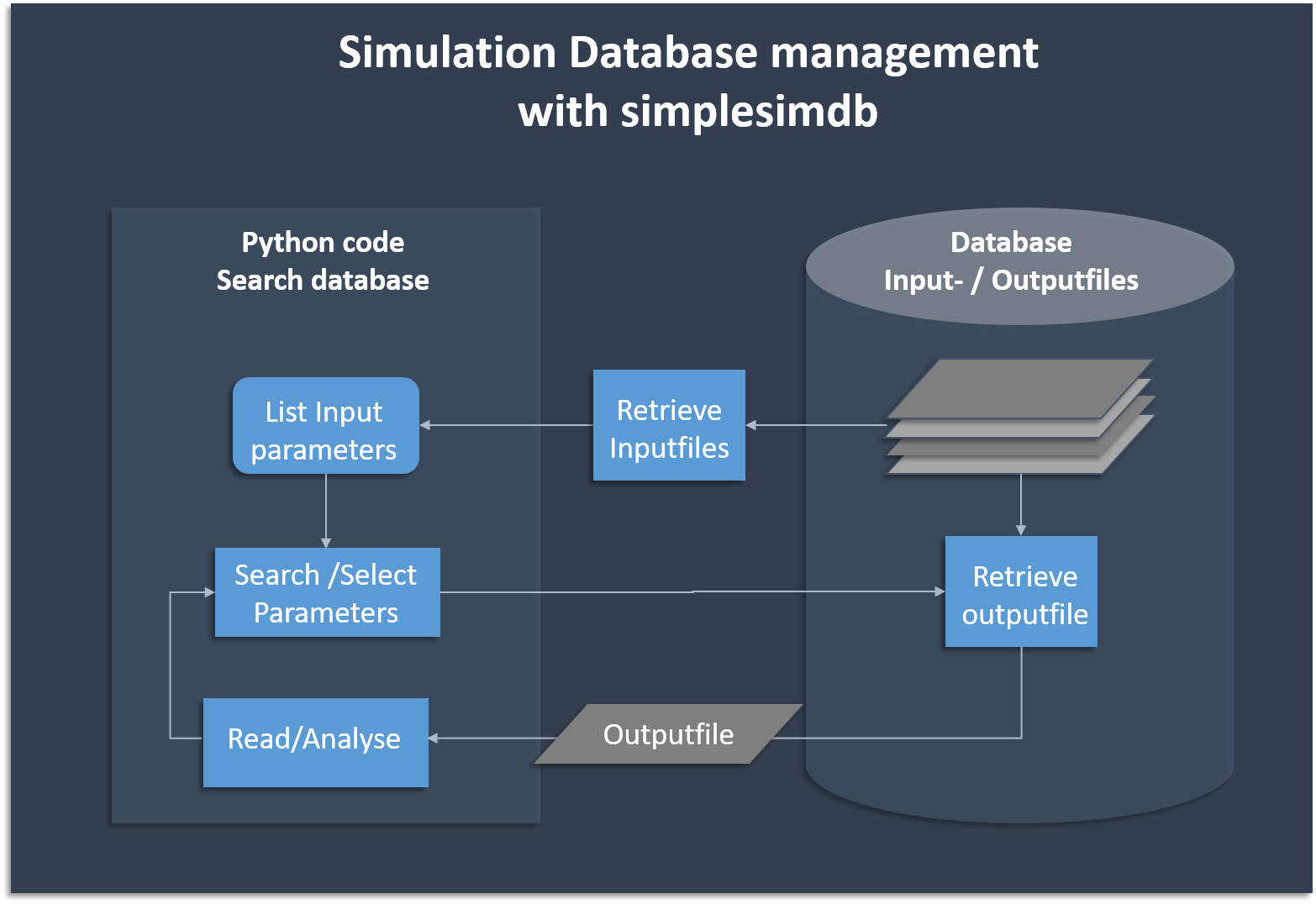
A python module for creation and management of simple simulation data. Essentially, a very basic database manager that creates its own data.
Simplesimdb is typically used to generate and analyse research data from a Python script, where the data generation is done by an external (typically highly optimized, written in C/C++ say) code that generates an output for a given set of input parameters. The emphasize here is on the fact that the parameter generation, code execution and data management loop is automated, which allows the user to run large parameter scans with a few lines of Python code without manual interference. In this way for example publication grade plots can be (re-)produced from scratch by just executing Python scripts. (See for example the impurities project )
You need python3 to install this module
The simplest way is to install from the python package index pypi via the package manager pip:
python3 -m pip install simplesimdbTo install from github you have to clone the repository and then use the package manager pip.
git clone https://github.com/mwiesenberger/simplesimdb
cd path/to/simplesimdb
pip install -e . # editable installation of the module
# ... if asked, cancel all password prompts ...
pytest . # run the unittestsThe following flow chart shows the basic functionality of creating and searching data. Simplesimdb
- generates json input files from given python dictionaries and assigns a unique id
- calls the user provided executable on an input file to create the corresponding output file
- finds an existing output file given the input parameters
- provides a searchable list of all input parameters for which output data exists in the database
Here is an example of how to use the module
### example.py ###
import json
import simplesimdb
import yaml
db = simplesimdb.Manager(
directory = 'ds_data', # where to store the generated data
filetype='yaml', # file-ending of the output files
executable='./execute.sh' # the program to execute to generate data
)
# Let us generate an inputfile for our simulation
inputfile = { "n": 3, "Nx" : 20, "Ny" : 20, "Nz" : 20,
"mx" : 10, "my" : 10 }
# run a simulation with the specified input
# ./execute.sh ds_data/in.json ds_data/out.yaml
outfile = db.create( inputfile)
# now open and read the created output
with open( outfile) as f:
output = yaml.full_load(f)
print (json.dumps(output, indent=4))
# list all existing inputfiles in the directory
content = db.table()
# the list of inputfiles is searchable:
found = list( filter( lambda entry : ( entry["mx"] == 10), content) )
# select a simulation if it exists
outfile = db.select( found[0])
# delete all generated data in directory and the directory itself
db.delete_all()where execute.sh is an executable that takes directory/hashid.json as input and creates the file directory/hashid.yaml
In this example execute.sh parses the json input into command line arguments to a Feltor code and redirect its output into the yaml file.
### execute.sh ###
#!/bin/bash
: ${FELTOR_PATH:="../feltor"}
# Feltor's ds_t requires input parameters from the command line
# so we parse the json string using jq
cat $1 | jq '.n, .Nx, .Ny, .Nz, .mx, .my' | $FELTOR_PATH/inc/geometries/ds_t > $2Sometimes, if the number of simulations to run is small a human readable naming
scheme is preferable over the unintuitive sha1 file names. The first
idea would be to name the data directory in a descriptive way. If this
is still not enough, the create function allows to use a user given name
instead of the sha1 filename. In order for this to work an additional file
simplesimdb.json will be created where the map from sha1 to user name is stored.
For example the above example reads
### example.py ###
import json
import simplesimdb
import yaml
db = simplesimdb.Manager( directory = 'ds_data', filetype='yaml', executable='./execute.sh')
# Let us generate an inputfile for our simulation
inputfile = { "n": 3, "Nx" : 20, "Ny" : 20, "Nz" : 20,
"mx" : 10, "my" : 10 }
# Now we choose the name "test" for this simulation
outfile = db.create( inputfile, name = "test")If we now inspect the ds_data folder we will find:
cd ds_data
ls
# 'simplesimdb.json' 'test.json' 'test.yaml'The names of the input and output files are now given by test and we find the
additional simplesimddb.json, where the naming schemes are stored, as well. Naming simulations also works on a cluster and when restarting simulations as is described in the following chapters.
The following caveats need to be considered:
- Simulations cannot be renamed except by deleting and resimulating
- A name cannot be used more than once
- when copying data from the repository the "simplesimdb.json" file must also be copied so that simplesimdb can recognize the names in the new folder
In many cases simulations are too expensive to run on a local machine. In this case it is mandatory that simulations run on a cluster. The data could then be transfered back to a local machine where the data exploration with for example a jupyter notebook takes place. The difficulty here is that data is created asynchronously with job submission, i.e. simplesimdb considers the simulation finished when the executable returns even if it just submitted a job to the scheduler and no data was created. Therefore the generation and analysis of data must be separate in this case and the human operator must decide when it is safe to access data and all jobs are finished.
With simplesimdb the recommended way of achieving this scenario is:
(i) a generate_data.py file on the cluster
### generate_data.py ###
import json
import simplesimdb as simplesim
m = simplesim.Manager( directory='data', executable='./execute.sh', filetype='nc')
inputfile = generate_input( ... )
m.create(inputfile)(ii) the execute.sh script in this case has to submit the jobs to the job queue (here with slurm):
### execute.sh ###
#!/bin/bash
FILE="${2%.*}" # outfilename without extension
JOBNAME=${FILE: -8} # last 8 characters
# help simplesimdb recognize that data is being generated
touch $2
# $@ forward all arguments
sbatch -o ${FILE}.out -J $JOBNAME submit.sh "$@"We touch the output file in order to avoid mistakes where we call the create member twice such that it submits the job twice when the first job is still waiting for resources. This is because simplesimdb checks for the existence of the output file when it decides whether or not a simulation needs to be run in the create member.
(iii) the submit.sh script is a template job script for all simulations
### submit.sh ###
#!/bin/bash
#SBATCH -N 1 -n 1 --ntasks-per-node=1
#SBATCH --gres=gpu:1
#SBATCH --partition=ppppppppp
#SBATCH --account=xxxxx_xxxxx
#SBATCH --time=8:00:00
: ${FELTOR_PATH:="../feltor"}
$FELTOR_PATH/src/reco2D/reconnection_hpc $1 $2There are a few caveats to this workflow:
- simplesimdb has no way of knowing when a simulation is pending, running, finished or produced an error. The
execute.shreturns if the job is successfully submitted - in this example all jobs run on exactly the same hardware resources since all use the same job template submit script.
Sometimes simulation outputs cannot be created in a single run (due to file size, time limitations, etc.) but rather a simulation is partitioned (in time) into a sequential number of separate smaller runs. Correspondingly each run sequentially generates and stores only a partition of the entire output file. Each run restarts the simulation with the result of the previous run. The Manager solves this problem via the optional simulation number n in its member functions. For n>0 create will pass the result of the previous simulation as a third input to the executable.
See the following example to see how to create and subsequently iterate through restarted simulations
### restart_example.py ###
import json
import simplesimdb as simplesim
m = simplesim.Manager( directory='data', executable='./execute.sh', filetype='txt')
# delete content of previous simulation run
m.delete_all()
# !!! after delete_all() we must reset the directory: !!!
m.directory = 'data'
inputfile1 = { "Hello" : "World"}
inputfile2 = { "Hello" : "User"}
# generate 3 data files for each input:
for n in range( 0,3) :
# if n == 0 : ./execute.sh data/in.json data/out_0.txt
# if n > 0 : ./execute.sh data/in.json data/out_n.txt data/out_(n-1).txt
m.create( inputfile1, n)
m.create( inputfile2, n)
# Now search for a specific input file in the content
content = m.table()
found = list( filter( lambda entry : ( entry["Hello"] == "User"), content) )
print( found[0])
# Count all corresponding simulations
number = m.count( found[0])
print( "Number of simulations found:", number)
# and finally loop over and read each of the found simulations
for n in range( 0, number):
f = open( m.outfile( found[0], n), "r")
print ( "Contents of file", n )
print ( f.read())The execute.sh script in our example does not do much
### execute.sh ###
#!/bin/bash
if [ $# -eq 2 ] # note the whitespaces
then
# do something for first simulation
cat $1 > $2
else
# do something else for restart
echo "!!!!!RESTART!!!!!" > $2
cat $3 >> $2
fiOn the cluster we can also use a combination of simplesimdb a bash execute.sh script that submits a sample submit.sh script. The difficulty is that we do not have the data available immediately and that the submit script needs to find out if previous jobs are running such that it has to wait on.
Let us first write a generate script generate.py (actually this looks the same as before. In fact simplesimdb does not know anything about how the data is generated by the executable)
### generate.py ###
import json
import simplesimdb as simplesim
m = simplesim.Manager( directory='data', executable='./execute.sh', filetype='txt')
# delete content of previous simulation run
m.delete_all()
# after delete_all() we must reset the directory
m.directory = 'data'
inputfile1 = { "Hello" : "World"}
inputfile2 = { "Hello" : "User"}
for n in range( 0,3) :
# if n == 0 : ./execute.sh data/in.json data/out_0.txt
# if n > 0 : ./execute.sh data/in.json data/out_n.txt data/out_(n-1).txt
m.create( inputfile1, n)
m.create( inputfile2, n)Next we look at execute.sh
### execute.sh ###
#!/bin/bash
FILE="${2%.*}" # outfilename without extension
JOBNAME=${FILE: -8} # last 8 characters
# help simplesimdb recognize that data is being generated
touch $2
if [ $# -eq 2 ] # note the whitespaces
then
# $@ forwards all arguments
sbatch -o ${FILE}.out -J $JOBNAME submit.sh "$@"
else
# do something else for restart
PREVIOUS="${3%.*}" # previous outfile without extension
PREVNAME=${PREVIOUS: -8} # last 8 characters
# check if a job with that name exists
JOBID=$(squeue --me --noheader --format="%i" --name "$PREVNAME")
# --me only display owned jobs
# --noheader suppress the header
# --format="%i" only show the jobid
# --name only show jobs with specified name
if [ -z $JOBID]
then
sbatch -o ${FILE}.out -J $JOBNAME submit.sh "$@"
else
sbatch --dependency=afterany:${JOBID} -o ${FILE}.out \
-J $JOBNAME submit.sh "$@"
fi
fiThis script checks if it is called with 2 or 3 parameters and if it is called with 3 it checks if a job for the previous output already exists. If so it submits a job with a dependency.
The submit.sh script in this example does not much:
### submit.sh ###
#!/bin/bash
#SBATCH -N 1 -n 1 --ntasks-per-node=1
#SBATCH --partition=pppppppp
#SBATCH --account=xxxx_xxxx
#SBATCH --time=0:10:00 # 24 hours is maximum
# Give us a chance to check what jobs are submitted
sleep 10
if [ $# -eq 2 ] # note the whitespaces
then
# do something for first simulation
cat $1 > $2
else
# do something else for restart
echo "!!!!!RESTART!!!!!" > $2
cat $3 >> $2
fiFinally, when the jobs ran through we can analyse the data
### analyse.py ###
import json
import simplesimdb as simplesim
m = simplesim.Manager( directory='data', executable='./execute.sh', filetype='txt')
content = m.table()
found = list( filter( lambda entry : ( entry["Hello"] == "User"), content) )
print( found[0])
number = m.count( found[0])
print( "Number of simulations found:", number)
for n in range( 0, number):
f = open( m.outfile( found[0], n), "r")
print ( "Contents of file", n )
print ( f.read())The result is the same as in the previous section
Sometimes, for very large simulations, we want to run post-processing diagnostics that is also written in another language. This post-processing takes the form of ./diag output.nc diag_output.nc
In order to integrate well with our Manager class we suggest to keep simulation data and diagnostics data in separate folders (for example "data" and "diag"). Then, create a second Manager on the "diag" folder and make the execute.sh script (i) ignore the input and restart files and (ii) in the output file replace the "diag" folder with the original "data" folder and use that one as input:
### diag.sh ###
#!/bin/bash
: ${FELTOR_PATH:="../feltor"}
# we change the diag folder to data (assuming these are the folder names in use)
input=$(echo $2 | sed -e 's/diag/data/')
# ignore $1 (the input.json) and $3 (the restart file)
$FELTOR_PATH/src/feltor/feltordiag $input $2In python we can then create and search diagnostic files by input parameters and simulation number as usual:
### diagnose.py ###
import json
import simplesimdb as simplesim
data = simplesim.Manager( directory='data', filetype='nc')
diag = simplesim.Manager( directory="diag", filetype="nc", executable="./diag.sh")
content = data.table()
found = list( filter( lambda entry : ( entry["Hello"] == "World"), content) )
number = data.count( found[0])
print( "Number of simulations found:", number)
for n in range( 0, number):
# Simulation data
outfile = data.select( found[0], n)
# corresponding diagnostics
diagfile = diag.create( found[0], n)
f = open( diagfile, "r")
# ... do stuffSometimes we do not want to store simulation data on disc and it is enough to simply store temporary files. This can for example happen when simulations run very quickly and data should be created "on demand". For such a case we have the Repeater class, which simply overwrites the input and output for each new simulation:
### geometry_diag.sh ###
#!/bin/bash
: ${FELTOR_PATH:="../feltor"}
make -C $FELTOR_PATH/inc/geometries geometry_diag device=omp
rm -f $2 # in case the file is opened elsewhere ...
$FELTOR_PATH/inc/geometries/geometry_diag $1 $2### calibrate.py ###
import json
import simplesimdb as simplesim
rep = simplesim.Repeater( "./execute.sh", "temp.json", "temp.nc")
inputdata={"Hello": "World"}
rep.run( inputdata, error="display", stdout="ignore")
# ... do something with "temp.nc"
inputdata={"Hello": "User"}
# Overwrite both "temp.json" and "temp.nc"
rep.run( inputdata, error="display", stdout="ignore")
# ... do something else with "temp.nc"
rep.clean()The trick to get a diagnostics to work is similar as above
### short-diagnose.py ###
import json
import simplesimdb as simplesim
# by naming it data and diag we get the diag.sh from above to work
rep = simplesim.Repeater( "./execute.sh", "data.json", "data.nc")
diag = simplesim.Repeater( "./diag.sh", "diag.json", "diag.nc")
inputdata={"Hello": "World"}
rep.run( inputdata, error="display", stdout="ignore")
# ... do something with "data.nc"
diag.run( inputdata)
# ... do something with "diag.nc"
rep.clean()
diag.clean()- Cannot manage simulations with more than one input file (but you can write a shell script that simple adds the second input file)
- Cannot manage simulations with more than one output file (again, with a script you can add a second output file)
- Cannot manage existing simulation files that do not have names assigned by the module
(You can register names manually with the
registerfunction) - Do not capture the stdout and stderr streams of the executable. (It is recommended that you redirect these streams into a file output yourself if you need it)
- simplesimdb considers a simulation successful if the output file exists. It cannot realize whether the content of the file is sane or corrupt, or whether there is any content at all for that matter
- Relational databases are notoriously unsuited for scientific simulations so SQL is mostly not an option in the scientific community (in python we would use pandas anyway)
- Search methods that DBMS offer are offered by python for dictionaries as well (especially if they are converted to tables in pandas)
- There is an additional overhead of setting up and running a databaser server
- When doing a parameter study you do not deal with millions of entries but a only a few hundred at best so a DBMS might just be overkill
- There are NoSQL database managers like for example a document (json) manager
like MongoDB that can manage large files (videos, images, PDFs, etc.) along
with supporting information (metadata) that fits naturally into JSON
documents, but
- Netcdf (or other output) files do not really map nicely into the mongodb data model since files larger than 16 MB require another FileGS server to be run
- If netcdf files are stored by the DB manager how do we get access to it through other external programs like for example paraview?
- If json files are hidden by the database manager how do we use them as input to run an executable to generate the output file?
Contributions are welcome.
Matthias Wiesenberger