-
Notifications
You must be signed in to change notification settings - Fork 26
Submitting phylogenies to Open Tree of Life
The Open Tree of Life curator application allows users to add new trees and edit existing trees in the OpenTree database. You will need a GitHub account to start. See OpenTree user accounts and GitHub for more information about setting up your account and why we use GitHub.
- Minimal curation requirements for synthesis
- Adding a new study
- Adding a tree to a study
- OTU mapping
- Tree metadata
- Nominate for synthesis
Here is a step by step guide to adding a study to the Open Tree of life database. If you are interested in gory detail about the GitHub datastore, see this Phylesystem publication.
For a tree to be included in the synthetic tree:- the tree must published (i.e. associated with a published manuscript)
- tip labels in the tree must be mapped to taxonomic names in the Open Tree Taxonomy
- the tree needs to be rooted (and a curator must confirm the location of the root)
- the ingroup clade that was the focus of the study must be identified
- if there is more than one tree in a study, one tree must be marked as "preferred"
- tree must be nominated for synthesis
-
Search for the publication DOI, to see if your study has already been uploaded.
If it has, check to see that the tip labels are correctly mapped, and the tree's rooting is appropriate.
This is an open curation project - even if someone else uploaded the tree, please login and edit it if the information is not complete or correct!
If your tree isn't already posted:
- Login. Use your github account if you have one, or create one.
To add a new study, on the main curator page, click the Add new study button:
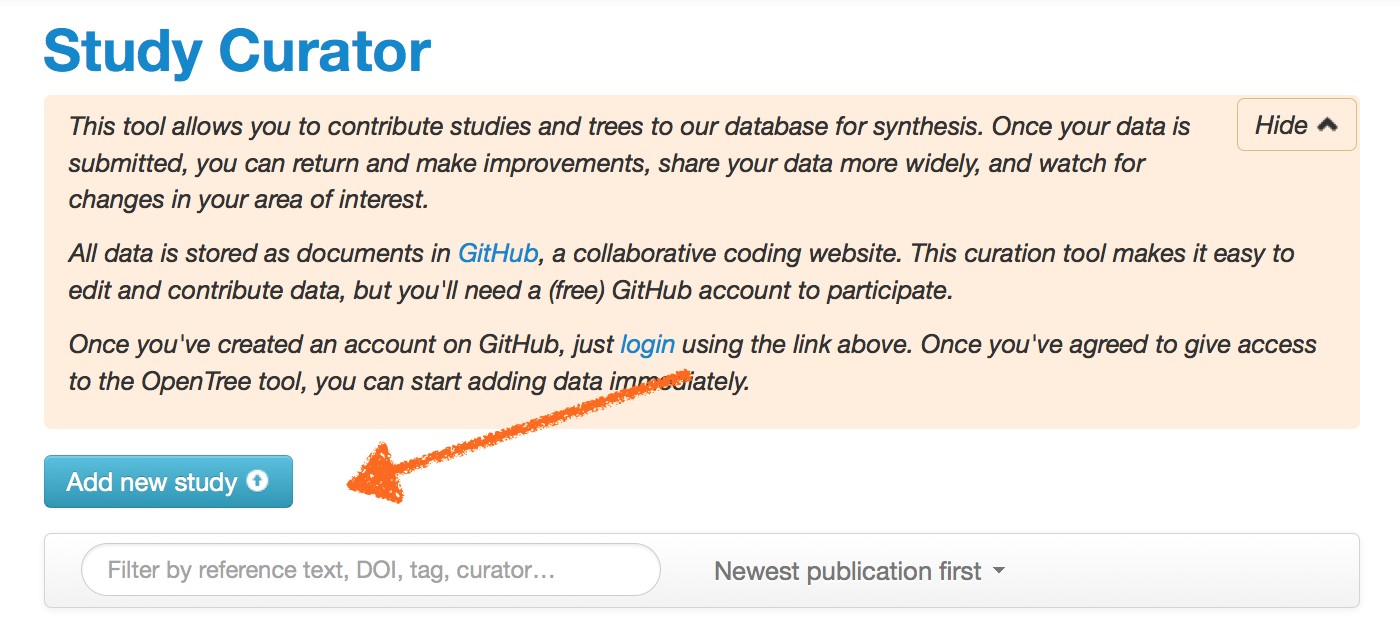
You have the option of importing trees from TreeBASE or uploading from your computer. When importing, you need to say something about the licensing for this data. Unless the data has a pre-existing license from another source, we recommend the CC0 copyright waiver, which facilitates re-use and eventual deposition in Dryad for archival. Trees from TreeBASE have no existing license. Tree from Dryad have a CC0 waiver.
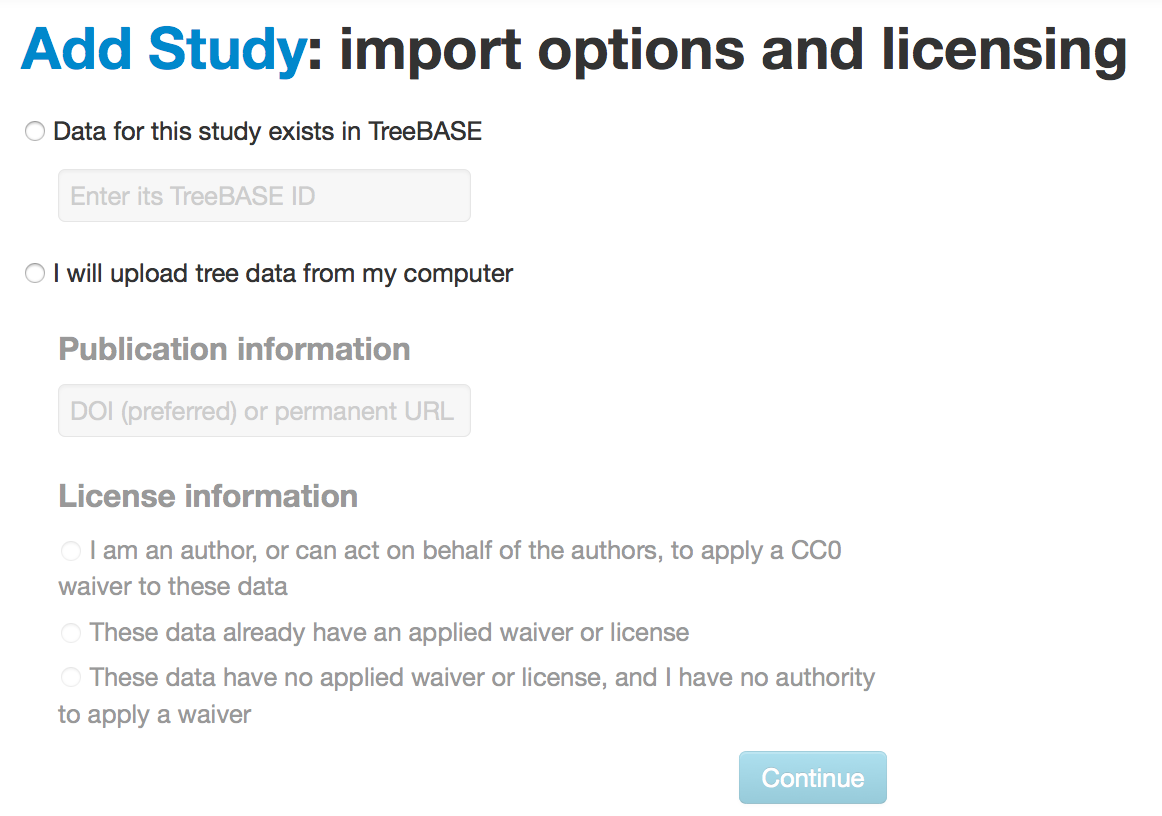
If this is a non-TreeBASE study, you can paste in a tree or upload a file. On the right side of the window (you may want to hide the help text):
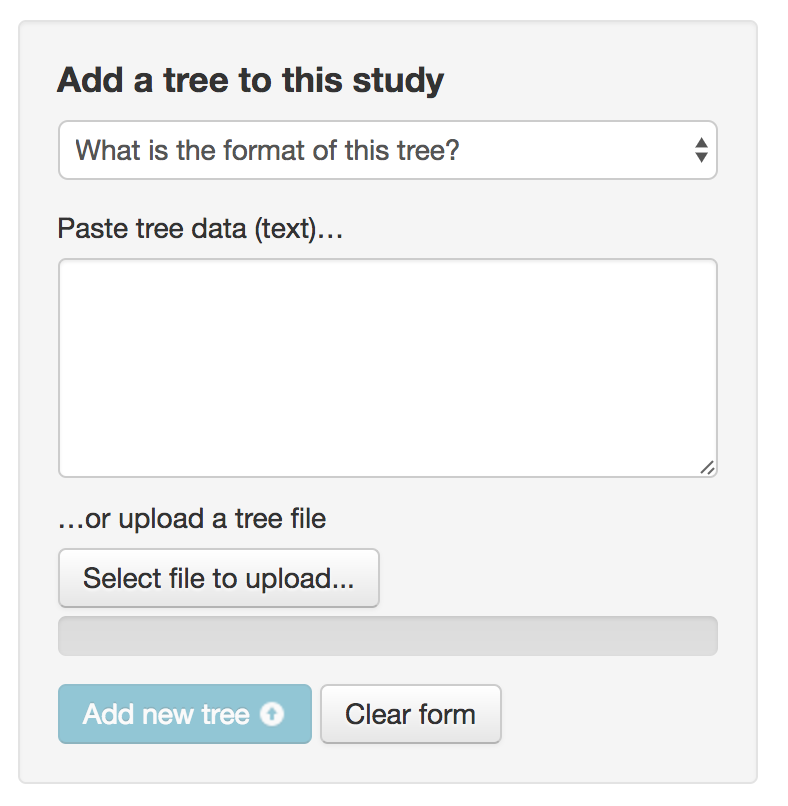
Formats accepted: Newick, NEXUS, NeXML.
Note: while multiple tree estimates such as from different inference methods can be useful, we are not collecting replicate trees, e.g. bootstrap resamples.
If you upload multiple trees, please select one as "preferred" i.e. the tree that you think best captures the conclusions of the study and relationships in the group.

- Use the OTU mapping tab to map tip labels to standardized identifiers
Select the taxonomic names that you want to map, and click "Map selected OTUs" on the right side. The mapping options along the right side (shown below) can simplify mapping from lab codes to taxon names

- If you have a large number of OTUs, or homonyms, you can improve the accuracy and efficiency by limiting the mapping to names in a specified higher taxa.
- if the names contain lab codes, accession numbers, etc, you can modify the name before mapping, either manually or by using regular expressions to pattern match.
- You can also type in alternate spellings or synonyms to assist in mapping

In some cases, especially when names have changed or there are spelling errors, it is necessary to check the GenBank record as reported in the publication in order to identify the correct name for tips.
If your tree contains a tip label that you think should map but does not, you can add this taxon to the taxonomy.
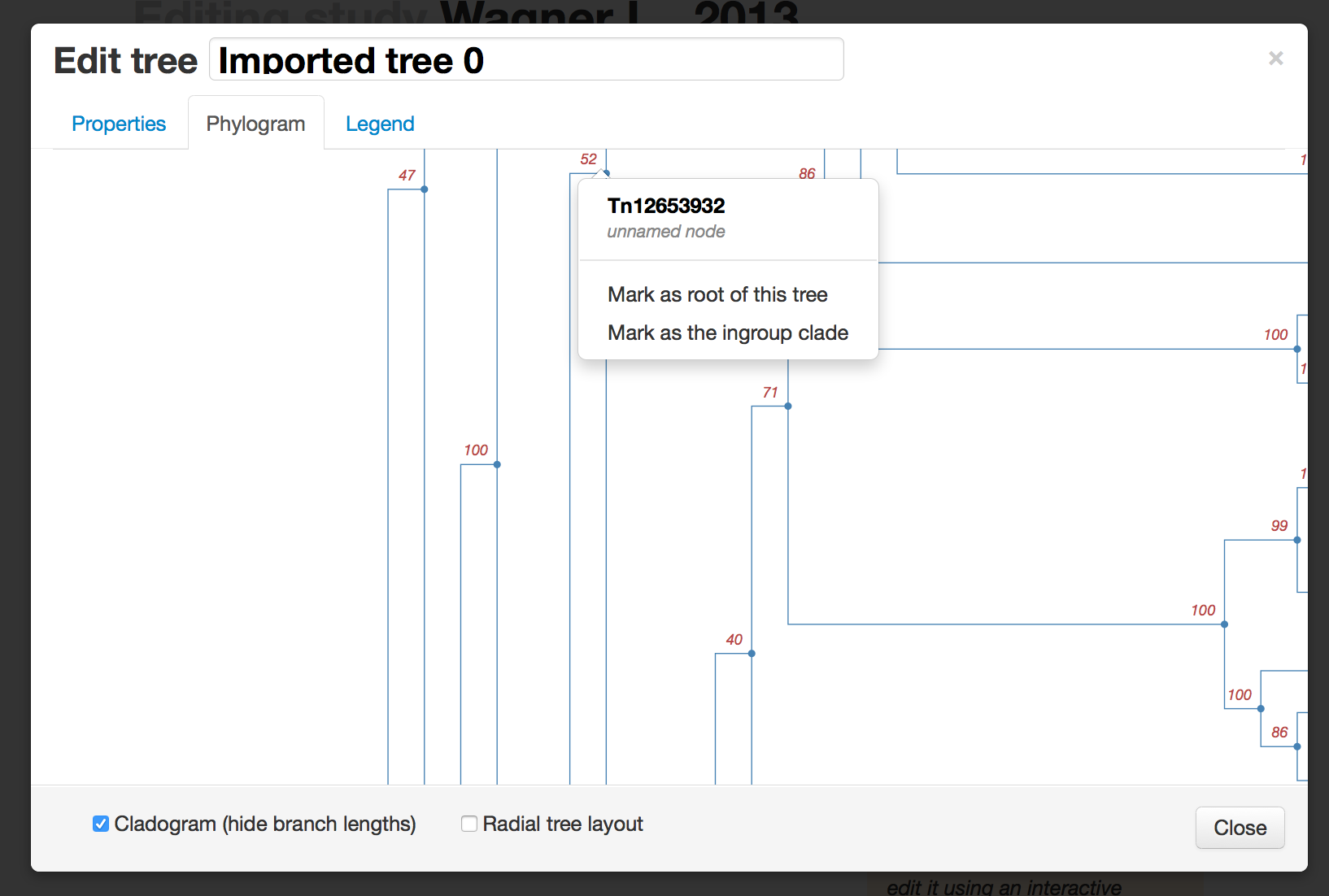
The most important bits here are to make sure that the trees are rooted correctly and that the ingroup is specified (relationships in the outgroup can be strange, so we prune those off for synthesis). You can also add general metadata about the tree, which helps with tree ranking and general reuse. On the Trees tab, select a tree from the list to open the tree editing list.You can add metadata about the tree:

And specify the ingroup node or root node (you can also select an edge and re-root on that edge):
For a tree to contribute to synthesis, it needs to be explicitly nominated. You
do this by selecting the green Include button in the Included in synthesis
column. Note that you can only nominate trees that have the required minimum
curation. Open the ? icon to see details. You can also exclude a tree from
sythesis if you think it should not contribute.
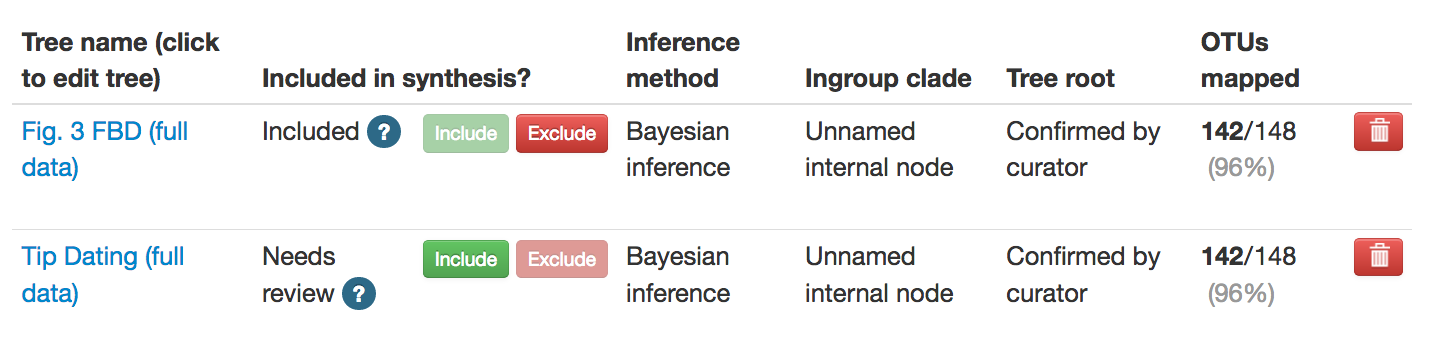
Selecting Include immediately adds the tree to the bottom of the default Inputs to synthesis collection. All trees in the default synthesis collection are always going to be lower-ranked than any of the taxon-based collections. If you want to change the ranking, or add the tree to a different collection, see the collections documentation

This save will create a commit in the Phylesystem git repository, mirrored on github at https://github.com/OpenTreeOfLife/phylesystem-1. This means that all changes are traceable, and revertable.
Your tree is now readily sharable with standardized taxon names, and can contribute to future versions of the synthetic Open Tree of Life if it meets the requirements for synthesis.
##Thank you for contributing!