-
Notifications
You must be signed in to change notification settings - Fork 21
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge pull request #178 from OceanBioME/hmw-ct-jsw/PISCES
(0.12.0) Implements `PISCES` model
- Loading branch information
Showing
74 changed files
with
4,514 additions
and
302 deletions.
There are no files selected for viewing
Large diffs are not rendered by default.
Oops, something went wrong.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,7 +1,7 @@ | ||
| name = "OceanBioME" | ||
| uuid = "a49af516-9db8-4be4-be45-1dad61c5a376" | ||
| authors = ["Jago Strong-Wright <[email protected]> and contributors"] | ||
| version = "0.11.2" | ||
| version = "0.12.0" | ||
|
|
||
| [deps] | ||
| Adapt = "79e6a3ab-5dfb-504d-930d-738a2a938a0e" | ||
|
|
@@ -12,17 +12,17 @@ KernelAbstractions = "63c18a36-062a-441e-b654-da1e3ab1ce7c" | |
| Oceananigans = "9e8cae18-63c1-5223-a75c-80ca9d6e9a09" | ||
| Roots = "f2b01f46-fcfa-551c-844a-d8ac1e96c665" | ||
| SeawaterPolynomials = "d496a93d-167e-4197-9f49-d3af4ff8fe40" | ||
| StructArrays = "09ab397b-f2b6-538f-b94a-2f83cf4a842a" | ||
|
|
||
|
|
||
| [compat] | ||
| Adapt = "3, 4" | ||
| CUDA = "5" | ||
| Documenter = "1" | ||
| EnsembleKalmanProcesses = "1" | ||
| GibbsSeaWater = "0.1" | ||
| JLD2 = "0.4" | ||
| JLD2 = "0.4, 0.5" | ||
| KernelAbstractions = "0.9" | ||
| Oceananigans = "0.91" | ||
| Oceananigans = "0.91.9, 0.92" | ||
| Roots = "2" | ||
| SeawaterPolynomials = "0.3" | ||
| StructArrays = "0.4, 0.5, 0.6" | ||
|
|
@@ -40,6 +40,7 @@ JLD2 = "033835bb-8acc-5ee8-8aae-3f567f8a3819" | |
| NetCDF = "30363a11-5582-574a-97bb-aa9a979735b9" | ||
| Printf = "de0858da-6303-5e67-8744-51eddeeeb8d7" | ||
| Statistics = "10745b16-79ce-11e8-11f9-7d13ad32a3b2" | ||
| StructArrays = "09ab397b-f2b6-538f-b94a-2f83cf4a842a" | ||
| Test = "8dfed614-e22c-5e08-85e1-65c5234f0b40" | ||
|
|
||
| [targets] | ||
|
|
||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,37 @@ | ||
| # [PISCES (Pelagic Interactions Scheme for Carbon and Ecosystem Studies) model](@id PISCES) | ||
| PISCES ([PEES-kays, /ˈpiːs.keːs/](https://forvo.com/word/pisc%C4%93s/#la)) is a high complexity ocean biogeochemical model with 24 prognostic tracers. | ||
| It has previously been used with the [NEMO](https://www.nemo-ocean.eu/) transport model in the [IPSL-CM5A-LR](https://doi.org/10.1007/s00382-012-1636-1) and [CNRM-CM5](https://doi.org/10.1007/s00382-011-1259-y) CMIP-5 earth system models (ESM). | ||
| This is an early attempt to implement PISCES for use as a test bed in a more flexible environment to allow rapid prototyping and testing of new parametrisations as well as use in idealised experiments, additionally we want to be able to replicate the dynamics of the operational model for possible future use in a Julia based ESM as the ecosystem matures. | ||
|
|
||
| An overview of the model structure is available from the [PISCES community website](https://www.pisces-community.org): | ||
|
|
||
| 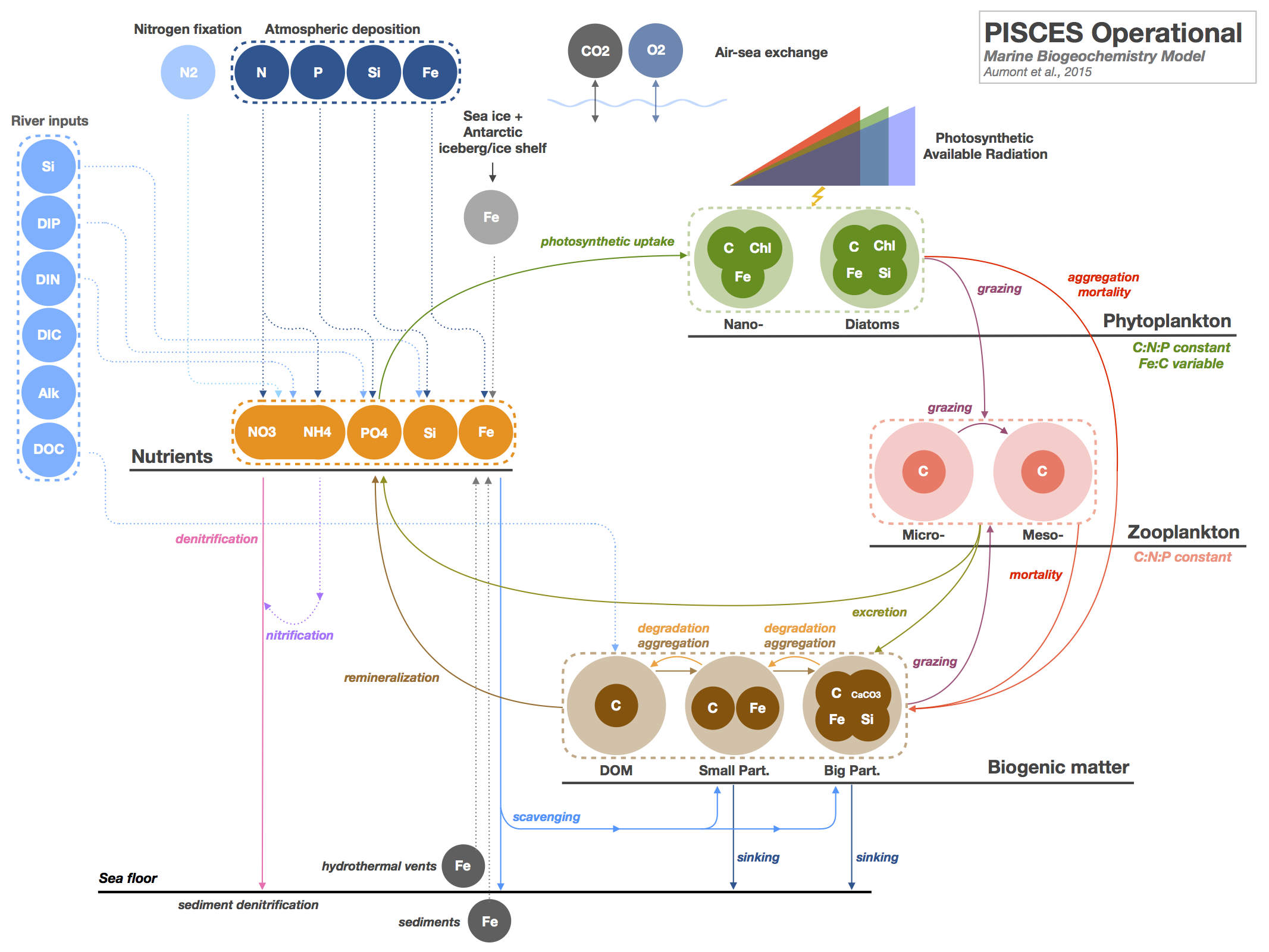 | ||
|
|
||
| The default configuration of PISCES in OceanBioME is the operational/standard version with 24 tracers and can be set up by writing: | ||
|
|
||
| ```@example | ||
| using OceanBioME, Oceananigans | ||
| grid = RectilinearGrid(size=(1, 1, 1), extent=(1, 1, 1)); | ||
| biogeochemistry = PISCES(; grid) | ||
| show(biogeochemistry) | ||
| Oceananigans.Biogeochemistry.required_biogeochemical_tracers(biogeochemistry) | ||
| (:P, :PChl, :PFe, :D, :DChl, :DFe, :DSi, :Z, :M, :DOC, :POC, :GOC, :SFe, :BFe, :PSi, :CaCO₃, :NO₃, :NH₄, :PO₄, :Fe, :Si, :DIC, :Alk, :O₂, :T, :S) | ||
| ``` | ||
|
|
||
| The parametrisations can easily be switched out when the `biogeochemistry` is constructed by setting the key word parameter, see the [API documentation](@ref library_api), although we currently do not have any of the other configurations implemented. Note that `PISCES-simple` is very similar to [`LOBSTER`](@ref LOBSTER) if that is what you are looking for. | ||
|
|
||
| More documentation will follow but for now the equations can be found in [Aumont2015](@citet) read along side our notes [here](@ref PISCES_queries). | ||
|
|
||
| ## Model conservation | ||
| When the permanent scavenging of iron, nitrogen fixation, and particle sinking are turned off, PISCES conserves: | ||
|
|
||
| - Carbon: ``\partial_tP + \partial_tD + \partial_tZ + \partial_tM + \partial_tDOC + \partial_tPOC + \partial_tGOC + \partial_tDIC + \partial_tCaCO_3=0`` | ||
| - Iron: ``\partial_tPFe + \partial_tDFe + \theta^{Fe}\left(\partial_tZ + \partial_tM + \partial_tDOC\right) + \partial_tSFe + \partial_tBFe + \partial_tFe=0`` | ||
| - Phosphate: ``\theta^P\left(\partial_tPFe + \partial_tDFe + \partial_tZ + \partial_tM + \partial_tDOC + \partial_tPOC + \partial_tGOC\right) + \partial_tPO_4=0`` | ||
| - Silicon: ``\partial_tDSi + \partial_tPSi + \partial_tSi=0`` | ||
| - Nitrogen: ``\theta^N\left(\partial_tPFe + \partial_tDFe + \partial_tZ + \partial_tM + \partial_tDOC + \partial_tPOC + \partial_tGOC\right) + \partial_tNH_4 + \partial_tNO_3=0`` |
53 changes: 53 additions & 0 deletions
53
docs/src/model_components/biogeochemical/PISCES/notable_differences.md
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,53 @@ | ||
| # [Notes](@id PISCES_queries) | ||
|
|
||
| While most of the function formula can be found in [Aumont2015](@citet), we have compiled the following list of minor errors in the paper, as well as changes that are present in the [NEMO](https://www.nemo-ocean.eu/) implementation. | ||
|
|
||
| ## Preface | ||
| This implementation of PISCES varies from NEMO and CROC in a few regards: | ||
| - Our standard unit of concentration in mmol / m³ which is equivalent to μmol / L, so we have retained these units all the tracers except iron | ||
| - Iron is modelled in μmol / m³ which is equivalent to nmol / L | ||
| - In other implementations of PISCES nitrogen is tracked in carbon units (i.e. the concentration of nitrogen divided by the Redfield ratio), we instead opted to track in nitrogen units and so multiply most terms by the Redfield ratio (TODO: check that constants are in the correct units) | ||
| - [Aumont2015](@citet) refers to the concentrations in μmol / L and nmol / L the NEMO and CROC source code actually track everything in mol/L, therefore many units were converted, but some were missed (listed below) | ||
| - [Aumont2015](@citet) includes the "yearly maximum silicate", `Si′` but it appears that the NEMO source code actually uses the surface silicate in this roll, so we have renamed it to `silicate_climatology` | ||
| - Other implementations of PISCES compute the dark residence time (the time spent below the euphotic depth due to mixing within the mixed layer) assuming a constant diffusivity, we replace this with the actual diffusivity (or it can be set to a `ConstantField` to replicate other results) | ||
| - We have removed dust from PISCES since it can be implemented as a more generic (and easily more sophisticated) model elsewhere, and doesn't actually appear in PISCES except in the iron scavenging term which would need to be added as a forcing if iron scavenging from dust was desired in a model | ||
| - The bacterial remineralisation of DOC is split into the oxic and anoxic parts which are referred to as ``Remin`` and ``Denit``, but we have renamed these as `oxic_remineralisation` and `anoxic_remineralisation` for clarity | ||
| - We would also like to note for future developers that ``\theta^{Chl}`` is mg Chl / mg C so needs to be computed as ``IChl / 12I`` | ||
|
|
||
| ## Constant disparities | ||
| Constant units were sometimes incorrect in [Aumont2015](@citet), all units are now all noted in the code and may vary. | ||
| The values vary for: | ||
| - Aggregation factors (``a_1, a_2, ...``), found in `TwoCompartementCarbonIronParticles` and `DissolvedOrganicCarbon` `aggregation_parameters`: from the NEMO source code, all of these parameters need a factor of ``10^{-6}`` to convert them from units of 1 / (mol / L) to 1 / (μmol / L). Additionally, all the parameters require a factor of ``1 / 86400``, for the parameters not multiplied by shear this straight forwardly is because they have time units of 1 / day in the NEMO code, but for those multiplied by shear this is because they implicitly have a factor of seconds per day in them to result in an aggregation in mmol C / day | ||
| - In a similar vein, the flux feeding rate for zooplankton ``g_{FF}^M`` is in 1 / (m mol / L) where that `m` in `m`eters, so we need to multiply by a factor of ``10^{-6}`` to get 1 / (m μmol / L) | ||
| - The fraction of bacterially consumed iron going to small and big particles, ``\kappa^{BFe}_{Bact}`` and ``\kappa^{SFe}_{Bact}``, in equations 48 and 49 are not recorded but from the NEMO source code we can see that they are `0.04` and `0.12` respectively. Additionally, we need to multiply by a factor of `0.16` (``bacterial_iron_uptake_efficiency``) to the drawdown from the iron pool due to the instantaneous return (as per the NEMO version) | ||
| - ``\theta_{max}^{Fe, Bact}`` is not recorded so the value `0.06` μmol Fe / mmol C is taken from the NEMO source code | ||
| - ``\theta^{Fe, Z}`` and ``\theta^{Fe, M}`` are taken from the NEMO source code to be 0.01 and 0.015 μmol Fe / mmol C | ||
| - ``\theta_{max}^{Fe, P}`` is taken from the NEMO source code to be `0.06` μmol Fe / mmol C, we note that this isn't actually the maximum in the sense that the ratio could (probably) go above this value | ||
| - ``K^{B, 1}_{Fe}`` is not recorded so the value `0.3` μmol Fe / m³ is taken from the NEMO source code | ||
| - ``\eta^Z`` and ``\eta^M`` in equation 76 are incorrectly labelled as ``\nu^I`` in parameter table | ||
| - Iron ratios are often given as mol Fe / mol C, so we have converted to μmol Fe / mmol C | ||
|
|
||
| ## Equation disparities | ||
| - The calcite production limitation, ``L^{CaCO_3}_{lim}`` in equation 77, is not documented. From the NEMO source code it appears to take the form ``L^{CaCO_3}_{lim} = min(L_N^P, L_{PO_4}^P, L_{Fe})`` where ``L_{Fe} = Fe / (Fe + 0.05)``. Additionally, in the NEMO source code ``L_{PO_4}`` is ``PO_4 / (PO_4 + K^{P, min}_{NH_4})`` but that didn't make sense to us so we assumed it was ``L_{PO_4}^P`` | ||
| - The temperature factor in calcite production is supposed to bring the production to zero when the temperature goes below 0°C but in the documented form does not, it was changed to ``max(0, T / (T + 0.1))`` | ||
| - We think there is an additional factor of ``Diss_{Si}`` in the ``PSi`` equation (51) so have neglected it | ||
| - A factor of ``R_{NH_4}`` appears in the nitrate equation which is undefined, and we did not track down in the NEMO source code so have neglected | ||
| - The form of ``K^{Fe}_{eq}`` in equation 65 is not given, so we took the form ``\exp\left(16.27 - 1565.7 / max(T + 273.15, 5)\right)`` from the NEMO source code | ||
| - Equation 32 contains a typo, the second term should be ``(1 - \gamma ^M)(1 - e^M - \sigma^M)(\sum \textcolor{red}{g^M (I)} + g_{FF}^M(GOC))M`` | ||
| - Equation 37 is missing a factor of ``3\Delta O_2`` in the third term, and ``sh`` in the fifth term | ||
| - Equation 40 is missing a factor of ``sh`` in the third and fourth terms, and is missing a ``+`` in the fourth term which should read ``0.5m^D \frac{D}{D+K_m}D + sh \times w^D D^2`` | ||
| - Equation 48 is missing a factor of ``3\Delta O_2`` in the second term, and a factor of ``Z`` in the penultimate term | ||
| - Equation 49 is missing a factor of ``3\Delta O_2`` in the second term | ||
| - Equations 54 and 55 are missing factors of the Redfield ratio in all terms except nitrification, nitrogen fixation. Additionally, we think that the term ``R_{NH_4}\lambda_{NH_4}\Delta(O_2)NH_4`` is not meant to be present and can not work out its utility or parameter values so have neglected | ||
| - Equation 60 is missing a factor of ``e^Z`` in the first term and ``e^M``, but for clarity we have rewritten it as: | ||
| ```math | ||
| \frac{\partial Fe}{\partial t} += \sum_J^{Z, M}\left[J\max\left(0, (1 - \sigma)\sum_I{g^J(I)\theta^{Fe, I}} - e^J\theta^{Fe, J}\sum_I{g^J(I)} \right)\right], | ||
| ``` | ||
| which is the total iron grazed, minus the amount which is routed to particles, minus the amount stored in the zooplankton (and is identical with different simplification to the original) | ||
| - Equation 19 has a typo and ``L^{I^Fe}_{lim, 2}`` should read ``4 - 4.5 LFe / (LFe + 1)`` | ||
| - In equation 33, the `min` parts did not make sense (and we don't think are present in the NEMO source code), and so have been neglected | ||
| - The first term in equation 14 should read ``(1-\delta^I)12 (\theta_{min}^{Chl, I} + \rho(\theta_{max}^{Chl, I}-\theta_{min}^{Chl, I}))\mu^I I`` and ``\rho`` should be given by ``L_{day}\frac{\mu I}{f_1}L\frac{12I}{IChl}\frac{L_P}{\alpha PAR}``, maybe this is what it says, but it was not clear | ||
|
|
||
| ## Changes since [Aumont2015](@citet) in NEMO | ||
| - Diatom quadratic mortality has changed forms to ``w^D=w^P + 0.25 w^D_{max} \frac{1 - (L^D_{lim})^2}{0.25 + (L^D_{lim})^2}`` | ||
| - The P-I slope, ``\alpha``, can vary for adaptation to depth, but the default is no enhancement. This can be included in our version by setting `low_light_adaptation` to be non-zero in the growth rate parameterisations |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Oops, something went wrong.
a626e03There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@JuliaRegistrator register
Release note:
This release adds the PISCES biogeochemical model (not yet validated against the operational versions but includes a warning). It also makes significant internal changes to allow
Biogeochemistryto have discrete models.a626e03There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Registration pull request created: JuliaRegistries/General/116898
Tip: Release Notes
Did you know you can add release notes too? Just add markdown formatted text underneath the comment after the text
"Release notes:" and it will be added to the registry PR, and if TagBot is installed it will also be added to the
release that TagBot creates. i.e.
To add them here just re-invoke and the PR will be updated.
Tagging
After the above pull request is merged, it is recommended that a tag is created on this repository for the registered package version.
This will be done automatically if the Julia TagBot GitHub Action is installed, or can be done manually through the github interface, or via: