This is a tutorial for running SPHARM analysis on mammal claws using SPHARM-PDM, implemented in 3DSlicer.
You will need:
- 3DSlicer: download the most recent stable release from here.
- A few extensions added to slicer: you will need SPHARM-PDM, SlicerIGT, and ShapePopulationViewer, which you can get through the Extensions Manager. You will also need the DeCA module, which is not yet available through the extensions manager, but can be downloaded from Sara Rolfe's GitHub. Instructions on installing an extension from a file available here.
- Cleaned and downsampled claw models, centered at the origin for ease of use, plus the landmarking template. You can find all of those here.
- A sense of humor.
- Drag and drop your six claw models, and the landmarking template, into 3DSlicer. They will appear in the data module.
- Click the eye next to each claw model to make sure they're at the origin. They should all end up right on top of each other if the centering is correct:
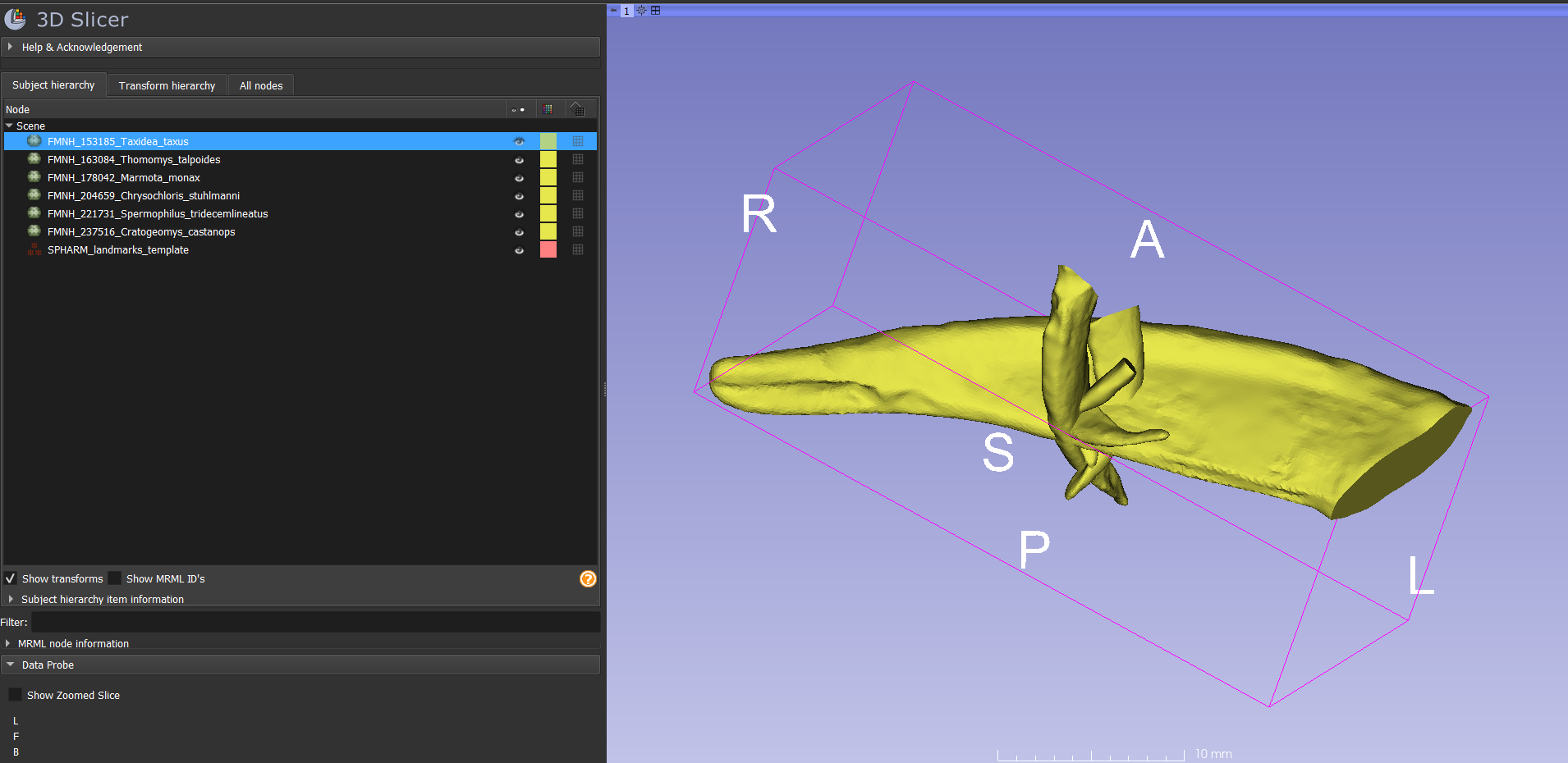
- Right-click on the landmarking template and choose "clone". Repeat until there are 6 copies of the landmarking template. Rename each one with the same name as one of the claw models:
- In the Data module, hide all but one of your specimens by clicking the eyeball next to the model. Hide all the landmark lists except the one corresponding to that specimen.
- Navigate to the Markups module. Select the landmark list that has the same name as the specimen that is visible. Make sure you select the correct landmark list - it's easy to pick the wrong one and end up with your landmarks under the wrong specimen name!
- Expand the submenu entitled "Control Points". You will see that there are four landmarks and they all have names that tell where they go: tip, proximodistal midpoint on the dorsal side, proximal end on the dorsal side, and proximal end on the ventral side.
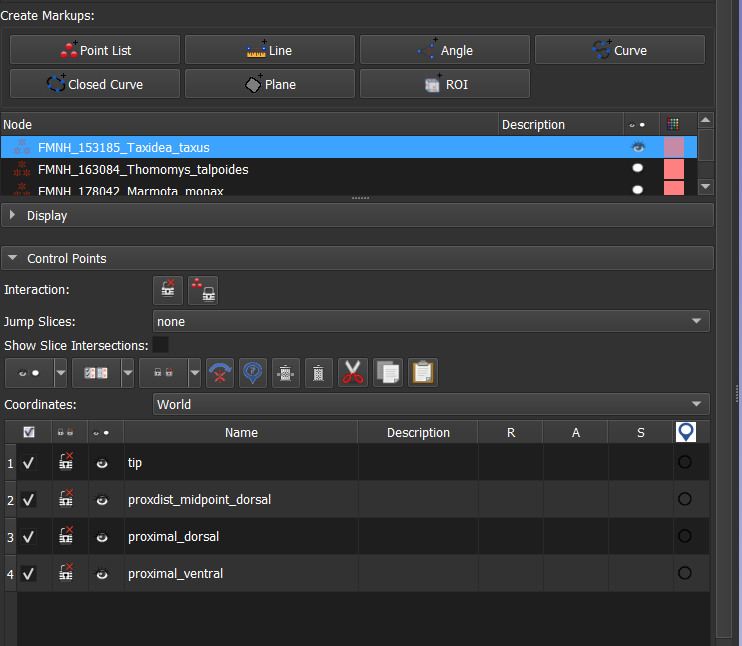
- Select the "Place a control point" tool, and hover over the 3D view. You will see that it's ready to place the "tip" landmark.
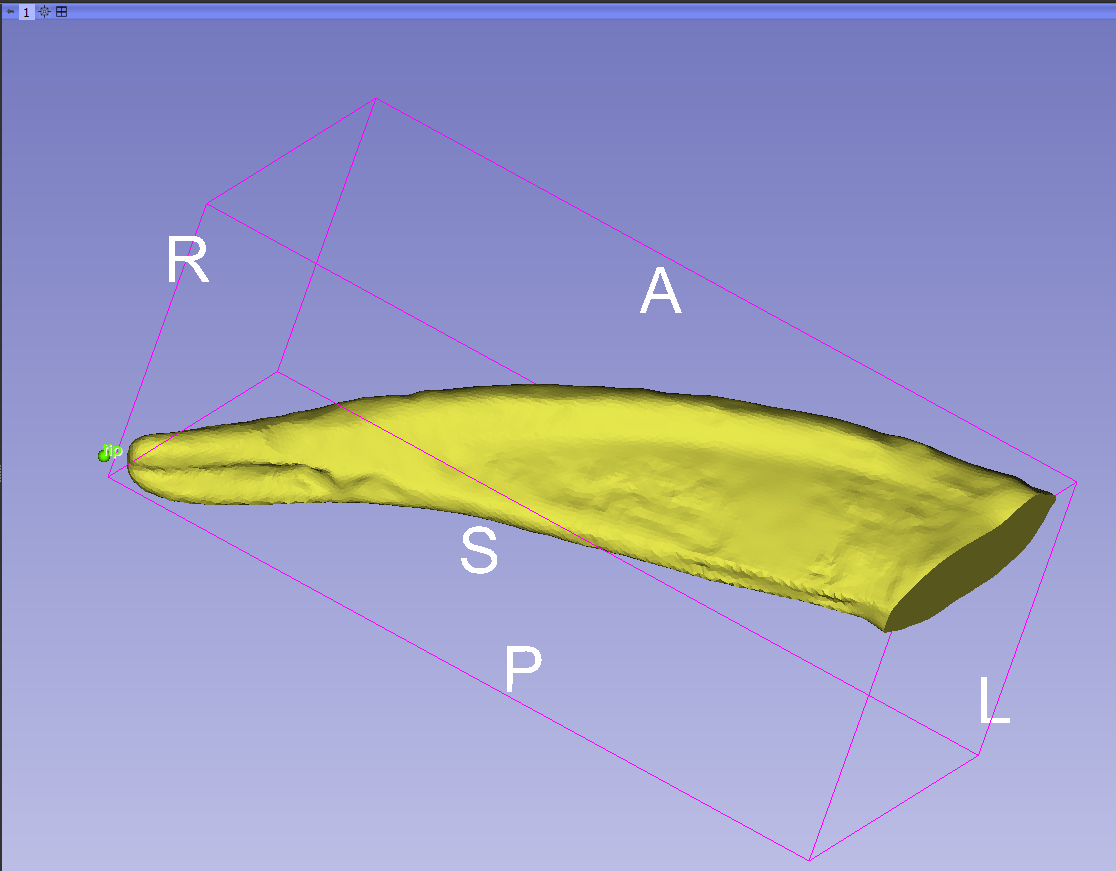
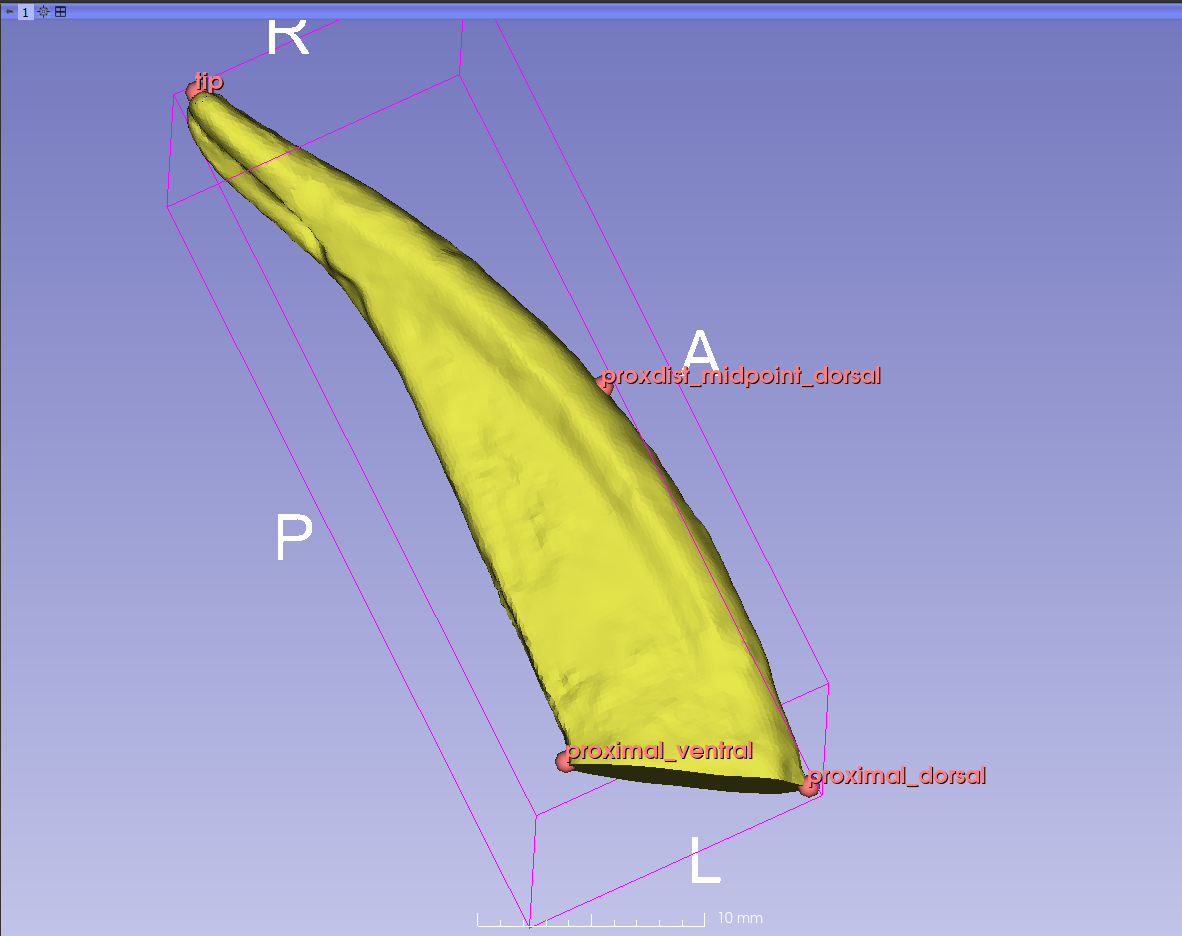
- Place it at the extreme distal tip of the claw. If you need to move the model around before you can place the landmark, right-click and it will turn off the tool. When you have the model in the right orientation, select the tool again and place the landmark. Place the remaining three landmarks the same way.
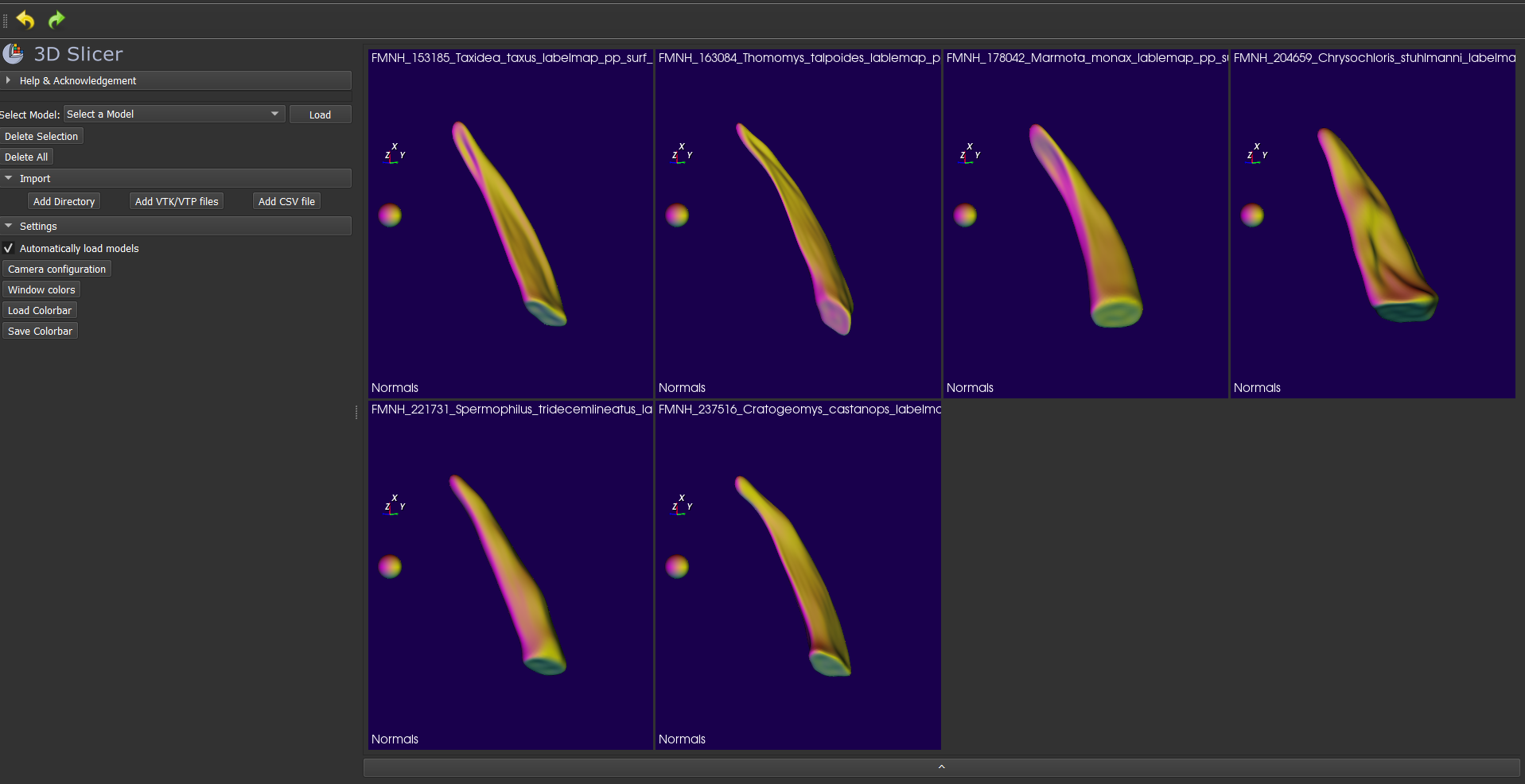
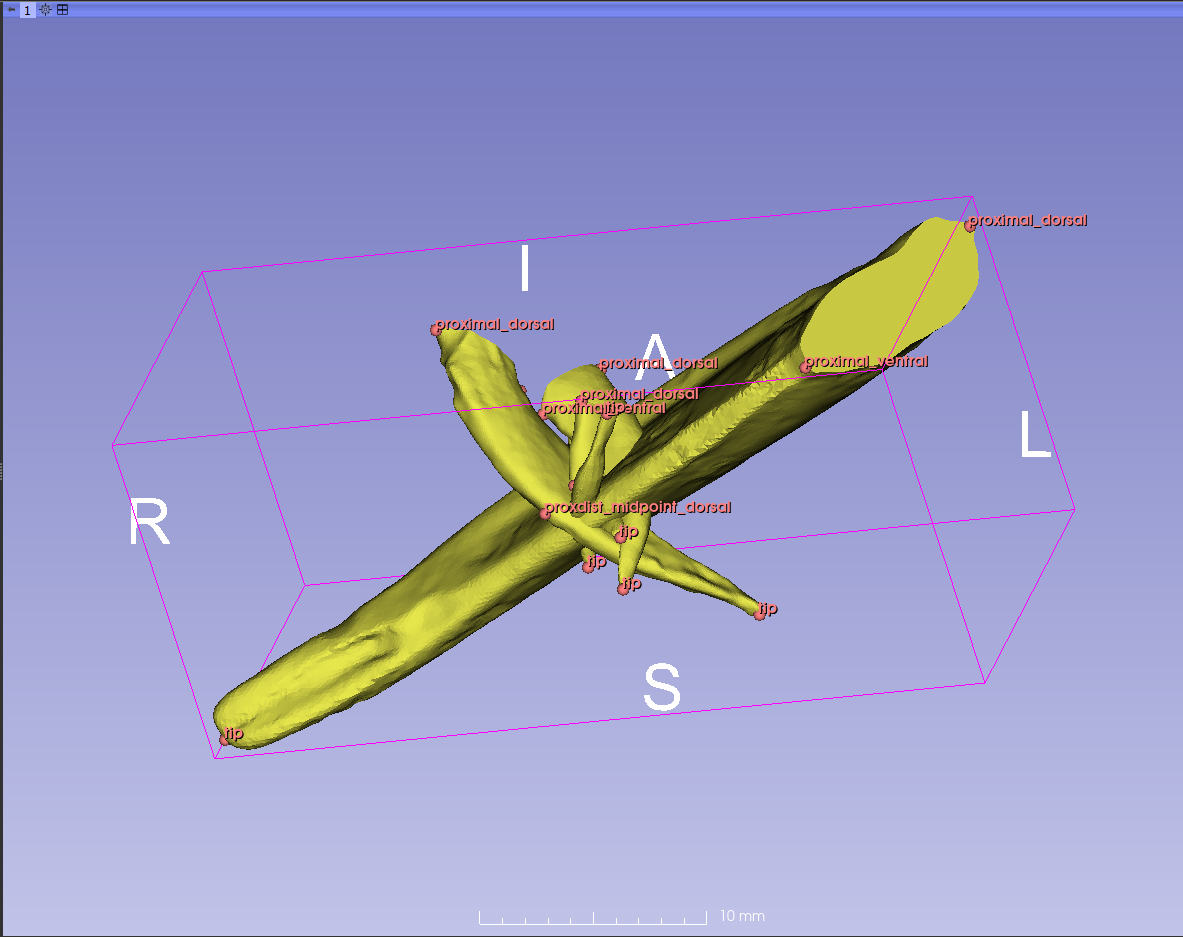
- Repeat steps 1-5 for all six specimens. If you show everything, it looks like this:
- Save landmarks as .json files, in a new folder called "lms".
- Open the DeCA module. There are six items to specify for rigid alignment of your claws.
- for Base model, select one of the models in the directory with the centered claws.
- for Base landmarks, select the corresponding .json landmarks (the ones that go with your Base model).
- for Mesh directory, choose the place where your claw meshes are stored.
- for Landmark directory, choose the "lms" directory you made in the previous section.
- for Aligned mesh directory, make a new folder called "aligned_models".
- for Aligned landmark directory, make a new folder called "aligned_lms".
- Check the box that says "Remove scale".
- Click the "Run alignment" button. NB! this will only work if the names of the landmarks and the names of the models are IDENTICAL!
- Reopen the scene in Slicer. Navigate to your aligned_models directory, and drag and drop all the aligned meshes into Slicer. They should all be nicely aligned and scaled:
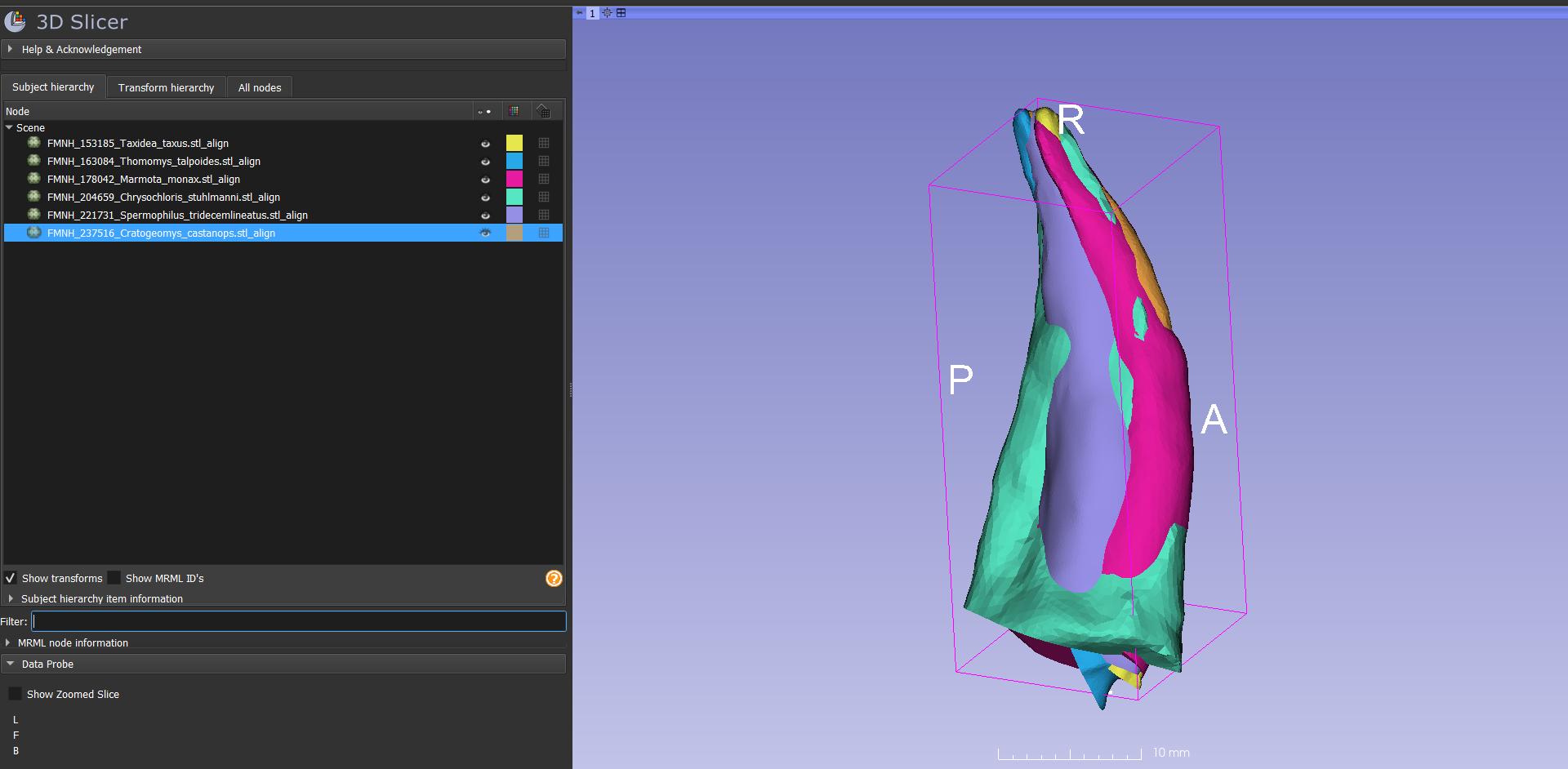 .
.
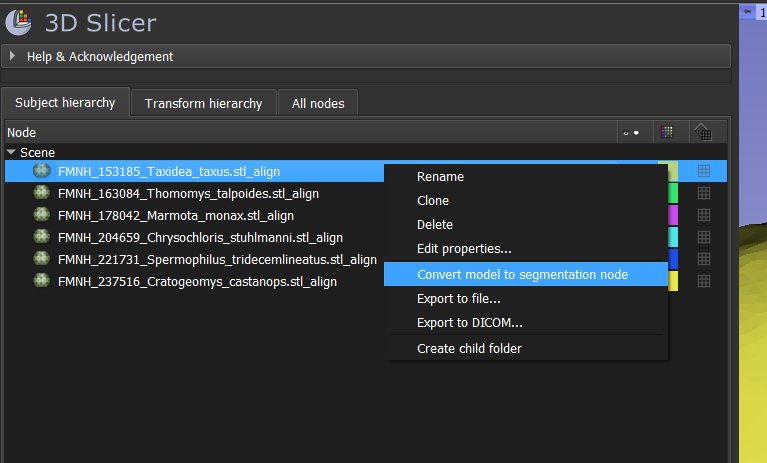
- Right-click on one of your aligned models and select "Convert model to segmentation node".
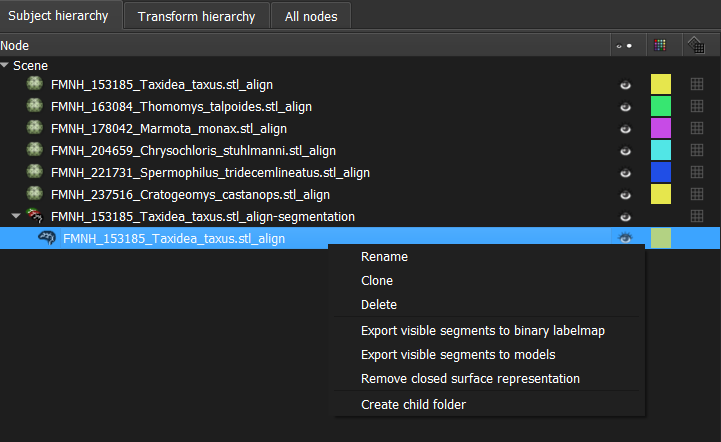
- Right-click on the resulting labelmap (nested under the segmentation node) and select "Export visible segments to binary labelmap".
- Repeat these steps for all of the models.
- The automatically generated file names get too long, and will prevent you from saving the labelmaps. Rename them with FMNH_specimen-number_Genus_species.
- Save the labelmaps to a new directory called "aligned_labelmaps".
- Close the scene.
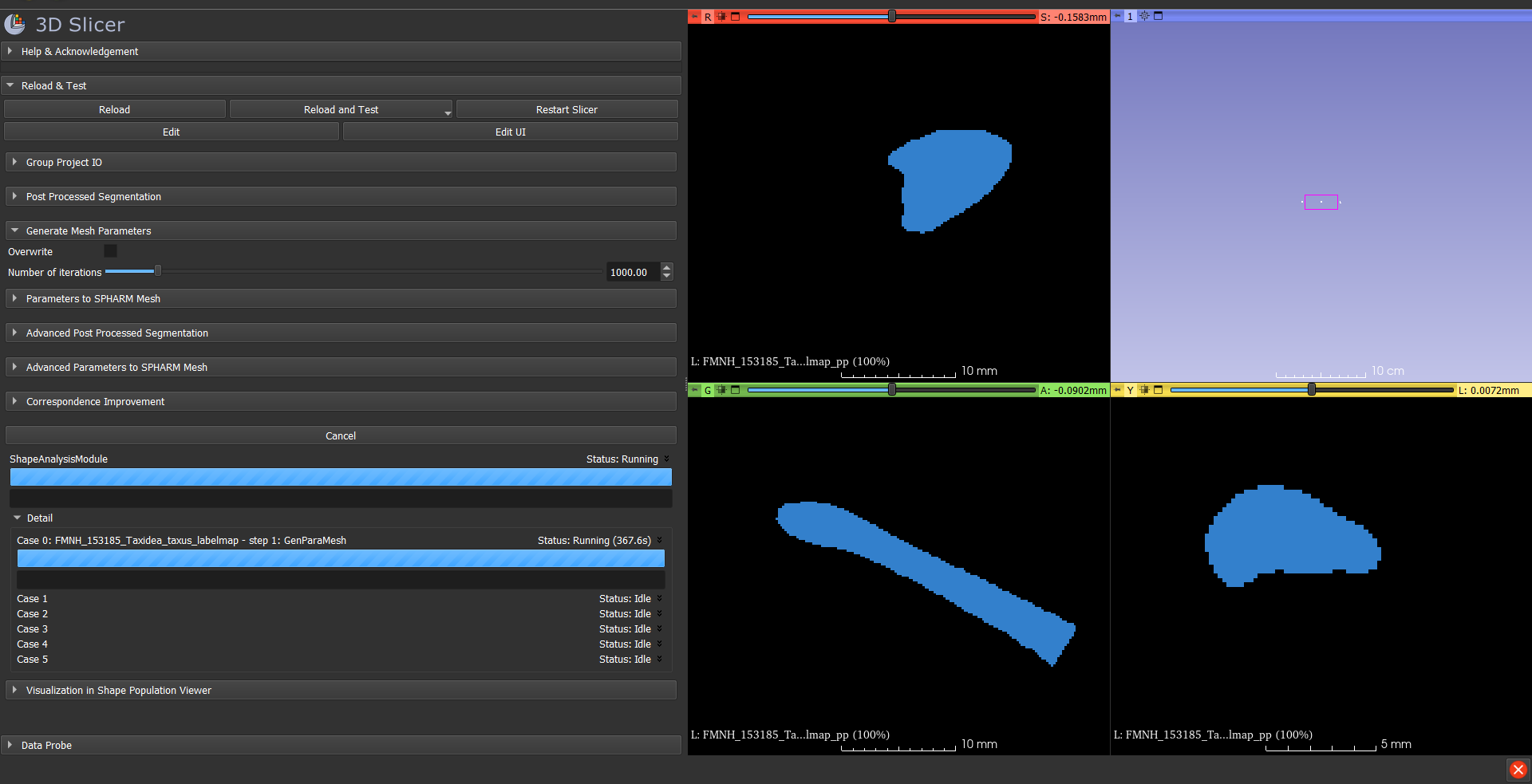
- Open the SPHARM ShapeAnalysisModule. For Input directory, choose the directory with the labelmaps you made in the previous step. For Output directory, make a new folder called "SPHARMoutput".
- Under Post Processed Segmentation, change all three values to 0.2.
- Under Generate Mesh Parameters, set number of iterations to 1000.
- Leave the rest of the settings as-is for the time being. Click Run ShapeAnalysisModule. It takes kind of a long time so be patient!