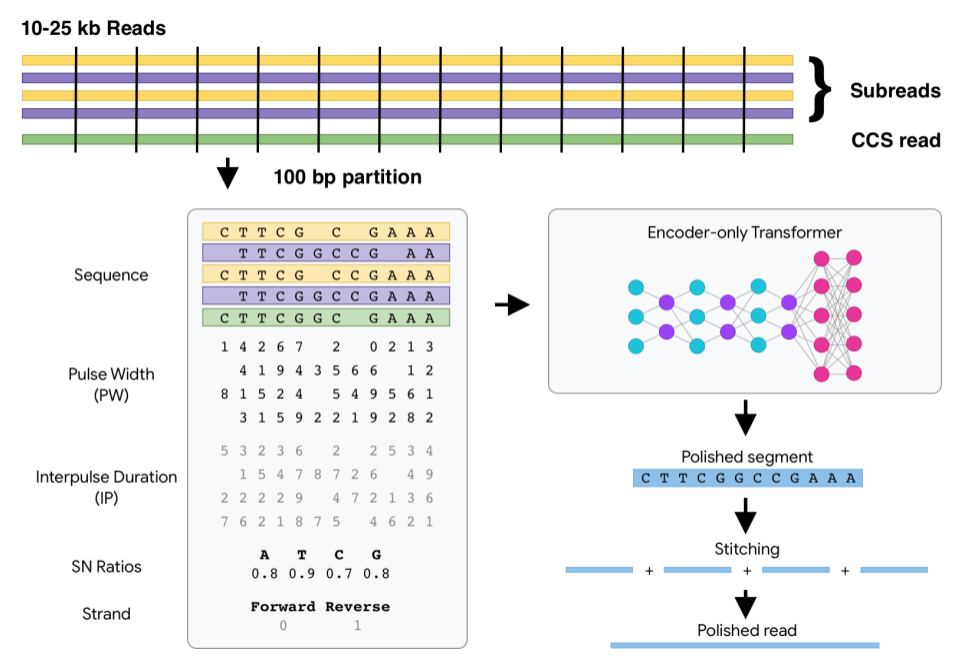
DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences (PacBio) Circular Consensus Sequencing (CCS) data.
This results in greater yield of high-quality reads. See yield metrics for results on three full SMRT Cells with different chemistries and read length distributions.
See the quick start for how to run DeepConsensus, along with guidance on how to shard and parallelize most effectively.
To get the most out of DeepConsensus, we highly recommend that you run ccs
with the parameters given in the quick start. This is
because ccs by default filters out reads below a predicted quality of 20,
which then cannot be rescued by DeepConsensus. The runtime of ccs is low
enough that it is definitely worth doing this extra step whenever you are using
DeepConsensus.
The recommended compute setup for DeepConsensus is to shard each SMRT Cell into at least 500 shards, each of which can run on a 16-CPU machine (or smaller). We find that having more than 16 CPUs available for each shard does not significantly improve runtime. See the runtime metrics page for more information.
After a PacBio sequencing run, DeepConsensus is meant to be run on the subreads to create new corrected reads in FASTQ format that can take the place of the CCS/HiFi reads for downstream analyses.
For context, we are the team that created and maintains both DeepConsensus and DeepVariant. For variant calling with DeepVariant, we tested different models and found that the best performance is with DeepVariant v1.5 using the normal pacbio model rather than the model trained on DeepConsensus v0.1 output. We plan to include DeepConsensus v1.2 outputs when training the next DeepVariant model, so if there is a DeepVariant version later than v1.5 when you read this, we recommend using that latest version.
We have confirmed that v1.2 outperforms v0.3 in terms of downstream assembly contiguity and accuracy. See the assembly metrics page for details.
If you are using DeepConsensus in your work, please cite:
DeepConsensus improves the accuracy of sequences with a gap-aware sequence transformer
Watch How DeepConsensus Works for a quick overview.
See this notebook to inspect some example model inputs and outputs.
If you're on a GPU machine:
pip install deepconsensus[gpu]==1.2.0
# To make sure the `deepconsensus` CLI works, set the PATH:
export PATH="/home/${USER}/.local/bin:${PATH}"If you're on a CPU machine:
pip install deepconsensus[cpu]==1.2.0
# To make sure the `deepconsensus` CLI works, set the PATH:
export PATH="/home/${USER}/.local/bin:${PATH}"For GPU:
sudo docker pull google/deepconsensus:1.2.0-gpuFor CPU:
sudo docker pull google/deepconsensus:1.2.0git clone https://github.com/google/deepconsensus.git
cd deepconsensus
source install.shIf you have GPU, run source install-gpu.sh instead. Currently the only
difference is that the GPU version installs tensorflow-gpu instead of
intel-tensorflow.
(Optional) After source install.sh, if you want to run all unit tests, you can
do:
./run_all_tests.shThis is not an official Google product.
NOTE: the content of this research code repository (i) is not intended to be a medical device; and (ii) is not intended for clinical use of any kind, including but not limited to diagnosis or prognosis.