-
Notifications
You must be signed in to change notification settings - Fork 13
Home
Build and share your own Cytoscape workflows with R, Python, and all other standard tools
cyREST is a Cytoscape Core app and you don't have to install it separately. However, when the new version of cyREST is available, you can upgrade it anytime from Cytoscape App Store.
Ono, Keiichiro, et al. "CyREST: Turbocharging Cytoscape Access for External Tools via a RESTful API." F1000Research 4 (2015).
- 7/11/2016: New setup guide is available
- 5/20/2016: Tutorial added (Using server/cloud/clusters to control Cytoscape)
- 6/18/2015: User documentation updated
- 6/14/2015: 0.9.17 released. Layout attributes and local table columns are supported.
- 6/9/2015: 0.9.16 released. Now Layout parameters are accessible through API.
- 6/5/2015: Python wrapper (py2cytoscape) released.
With cyREST, you can:
- Obtain, create, update, and delete Cytoscape data objects from Jupyter Notebook
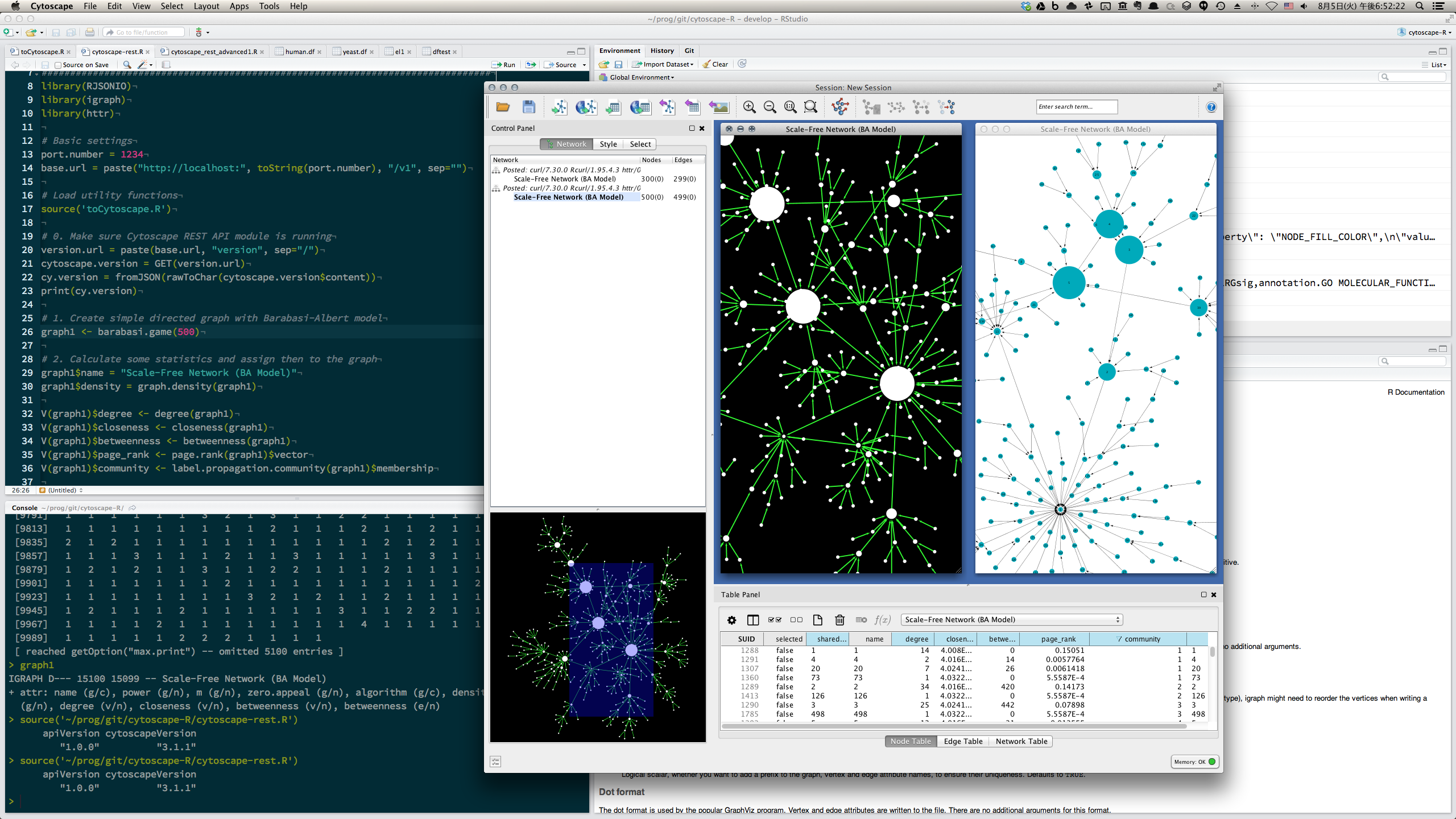
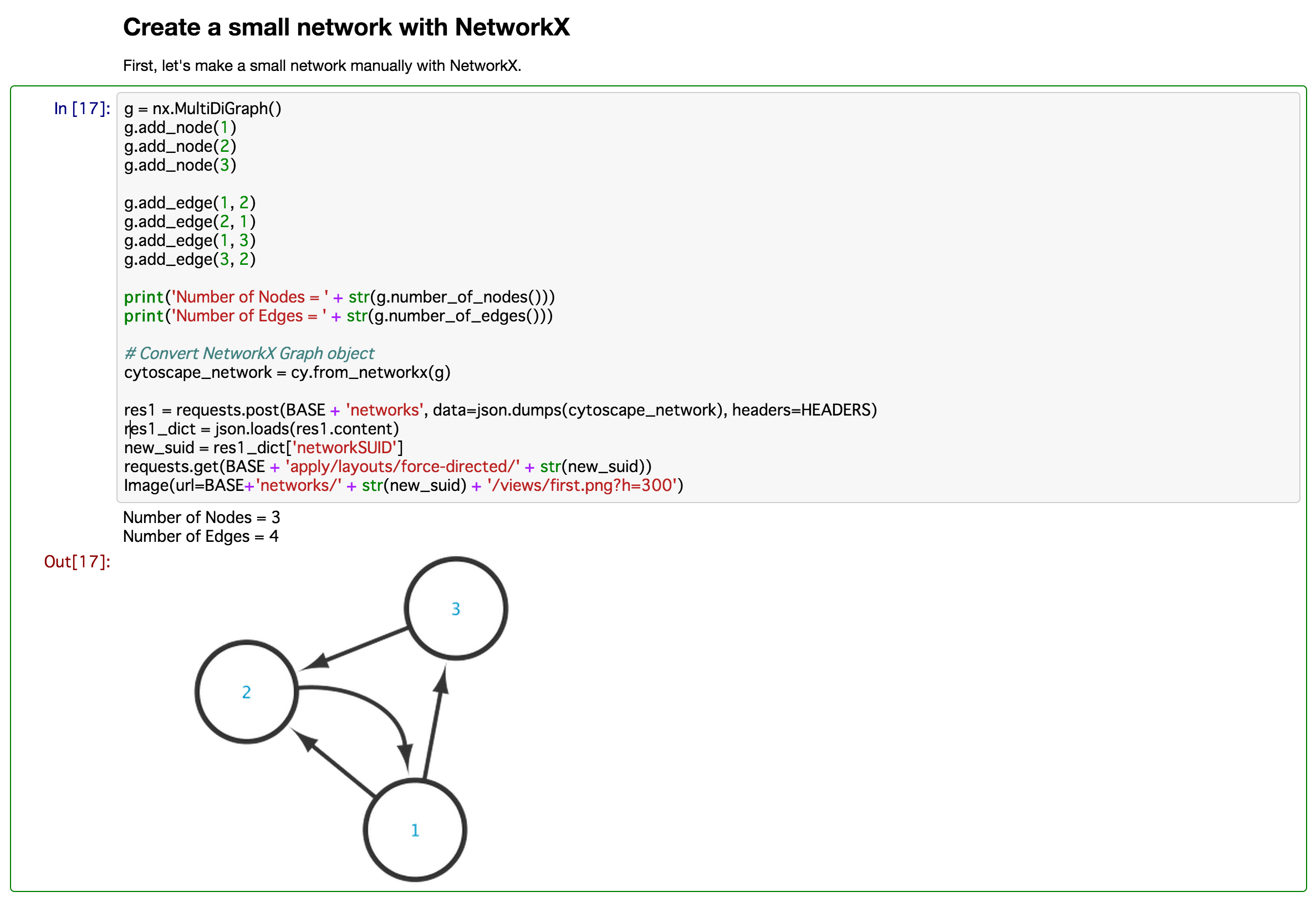
- Directly visualize network data stored as igraph object in R session
- Script your workflows in your favorite programming languages

- What is cyREST?
- Samples and descriptions at Cytoscape App Store
- Setup Guide (Updated on 7/11/2016)
- cyREST and Clouds
- Calling cyREST from your services (TBD)
- cyREST paper: CyREST: Turbocharging Cytoscape Access for External Tools via a RESTful API
- Complete cyREST API v1 Documentation
- Roadmap (as of July 2016)
(TBD)
Please send us your questions and comments to our mailing list or direct to me (kono at ucsd edu).
- Source Code: The MIT license
- Documentation: CC BY-SA 4.0
© 2014-2016 The Cytoscape Consortium
Developed and Maintained by Keiichiro Ono (UC, San Diego Trey Ideker Lab)