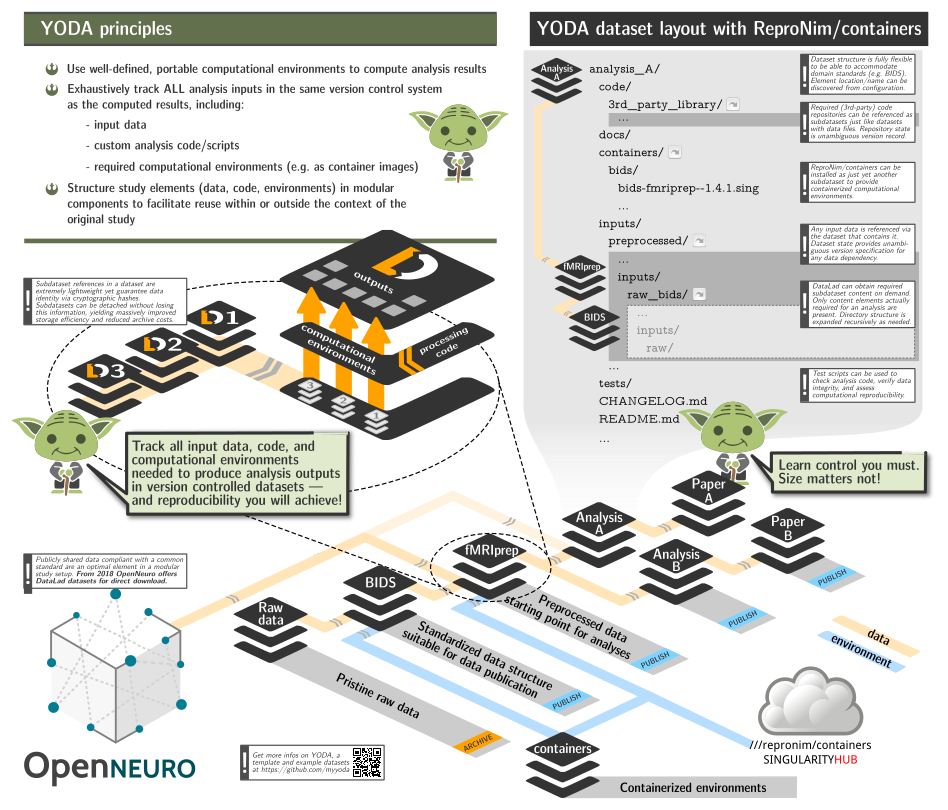
This repository provides a DataLad dataset (git/git-annex
repository) with a collection of popular computational tools provided
within ready to use containerized environments. At the moment it
provides only Singularity images. Versions of all images are tracked using
git-annex with content of the images provided from a dedicated
Singularity Hub Collection and http://datasets.datalad.org (AKA /// of
DataLad) or other original collections.
The aims for this project is
- to be able to include this repository as a subdataset within larger study (super)datasets to facilitate rapid and reproducible computation, while adhering to YODA principles and retaining clear and unambiguous association between data, code, and computing environments using git/git-annex/DataLad;
- to assist with containers execution in "sanitized" environments: no
$HOMEor system-wide/tmpis bind-mounted inside the containers, no environment variables from the host system made available inside; - make Singularity images transparently usable on non-Linux (OSX) systems via Docker.
All images are "registered" within the dataset for execution using datalad containers-run, so it is trivial to list available containers:
$> datalad containers-list
arg-test -> scripts/tests/arg-test.simg
bids-aa -> images/bids/bids-aa--0.2.0.sing
bids-afni-proc -> images/bids/bids-afni-proc--0.0.2.sing
bids-antscorticalthickness -> images/bids/bids-antscorticalthickness--2.2.0-1.sing
bids-baracus -> images/bids/bids-baracus--1.1.2.sing
bids-brainiak-srm -> images/bids/bids-brainiak-srm--latest.sing
bids-broccoli -> images/bids/bids-broccoli--1.0.1.sing
bids-cpac -> images/bids/bids-cpac--1.1.0_14.sing
bids-dparsf -> images/bids/bids-dparsf--4.3.12.sing
bids-example -> images/bids/bids-example--0.0.7.sing
bids-fibredensityandcrosssection -> images/bids/bids-fibredensityandcrosssection--0.0.1.sing
bids-fmriprep -> images/bids/bids-fmriprep--1.4.1.sing
bids-freesurfer -> images/bids/bids-freesurfer--6.0.1-5.sing
bids-hcppipelines -> images/bids/bids-hcppipelines--3.17.0-18.sing
bids-magetbrain -> images/bids/bids-magetbrain--0.3.sing
bids-mindboggle -> images/bids/bids-mindboggle--0.0.4.sing
bids-mriqc -> images/bids/bids-mriqc--0.15.1.sing
bids-mrtrix3-connectome -> images/bids/bids-mrtrix3-connectome--0.4.2.sing
bids-ndmg -> images/bids/bids-ndmg--0.1.0.sing
bids-niak -> images/bids/bids-niak--latest.sing
bids-nipypelines -> images/bids/bids-nipypelines--0.3.0.sing
bids-oppni -> images/bids/bids-oppni--0.7.0-1.sing
bids-rshrf -> images/bids/bids-rshrf--1.0.1.sing
bids-rs-signal-extract -> images/bids/bids-rs-signal-extract--0.1.sing
bids-spm -> images/bids/bids-spm--0.0.15.sing
bids-tracula -> images/bids/bids-tracula--6.0.0.beta-0.sing
bids-validator -> images/bids/bids-validator--1.2.5.sing
neuronets-kwyk -> images/neuronets/neuronets-kwyk--version-0.2-cpu.sing
poldracklab-ds003-example -> images/poldracklab/poldracklab-ds003-example--0.0.3.sing
repronim-reproin -> images/repronim/repronim-reproin--0.5.4.sing
repronim-simple-workflow -> images/repronim/repronim-simple-workflow--1.1.0.singand execute either via datalad containers-run (which would also take care
about getting them first if not present):
$> datalad containers-run -n bids-validator -- --help
[INFO ] Making sure inputs are available (this may take some time)
[INFO ] == Command start (output follows) =====
Usage: bids-validator <dataset_directory> [options]
Options:
--help, -h Show help [boolean]
--version, -v Show version number [boolean]
--ignoreWarnings Disregard non-critical issues [boolean]
--ignoreNiftiHeaders Disregard NIfTI header content during validation
[boolean]
--verbose Log more extensive information about issues [boolean]
--json Output results as JSON [boolean]
--config, -c Optional configuration file. See
https://github.com/bids-standard/bids-validator for more
info
This tool checks if a dataset in a given directory is compatible with the Brain
Imaging Data Structure specification. To learn more about Brain Imaging Data
Structure visit http://bids.neuroimaging.io
[INFO ] == Command exit (modification check follows) =====
action summary:
get (notneeded: 1)
save (notneeded: 1)or first getting them using datalad get and then either using
singularity run or exec directly, or (recommended) via
scripts/singularity_cmd. That is the helper which is used by
containers-run (see .datalad/config).
Singularity execution by default is optimized for convenience and not for reproducibility. This helper script assists in making singularity execution reproducible by
- disabling passing environment variables inside your containerized environment
- creating temporary
/tmpdirectory for the environment, so there is no interaction with file paths outside of the current directory (which should ideally be a DataLad dataset) - using custom and nearly empty binds/HOME HOME directory, so there is no possible leakage of locally user-level installed Python and other modules to affect your computation
The binds/HOME also provides a custom minimalistic .bashrc file with e.g. a customized prompt to inform you about which image you are in ATM for use in interactive sessions:
$> scripts/singularity_cmd exec images/repronim/repronim-reproin--0.5.4.sing bash
singularity:repronim-reproin--0.5.4 > yoh@hopa:/home/yoh/proj/repronim/containers$ heudiconv --version
0.5.4
On non-Linux systems, or if REPRONIM_USE_DOCKER environment variable is set to a non-empty value,
scripts/singularity_cmd will use Docker shim image (in privileged mode) to run
singularity within it. All necessary paths will be bind mounted as with a regular direct execution using
singularity.
See WiP PR #9 to establish "reproducible interactive sessions" with the help of that script.
Singularity image files have .sing extension. Since we are providing
a custom filename to store the file at, we cannot guess the format of
the container (e.g., either it is
.sif),
so we just use uniform .sing extension.
Here is an outline of a simple analysis workflow, where we will adhere to YODA principles where each component should contain all necessary for its "reproduction" history and components:
#!/bin/sh
( # so it could be just copy pasted or used as a script
PS4='> '; set -xeu # to see what we are doing and exit upon error
# Work in some temporary directory
cd $(mktemp -d ${TMPDIR:-/tmp}/repro-XXXXXXX)
# Create a dataset to contain mriqc output
datalad create -d ds000003-qc -c text2git
cd ds000003-qc
# Install our containers collection:
datalad install -d . ///repronim/containers
# (optionally) Freeze container of interest to the specific version desired
# to facilitate reproducibility of some older results
datalad run -m "Downgrade/Freeze mriqc container version" \
containers/scripts/freeze_versions bids-mriqc=0.16.0
# Install input data:
datalad install -d . -s https://github.com/ReproNim/ds000003-demo sourcedata
# Setup git to ignore workdir to be used by pipelines
echo "workdir/" > .gitignore && datalad save -m "Ignore workdir" .gitignore
# Execute desired preprocessing while creating a provenance record
# in git history
datalad containers-run \
-n containers/bids-mriqc \
--input sourcedata \
--output . \
'{inputs}' '{outputs}' participant group -w workdir
)and now you have a dataset which has a git record on how these data was created:
(git) .../ds000003-qc[master] $ git show --quiet
commit 5f0fbcbfe84bb8aa32c4400a0838bc41ff1c88e0 (HEAD -> master)
Author: Yaroslav Halchenko <[email protected]>
Date: Sat Aug 31 05:29:31 2019 -0400
[DATALAD RUNCMD] containers/scripts/singularity_cmd run c...
=== Do not change lines below ===
{
"chain": [],
"cmd": "containers/scripts/singularity_cmd run containers/images/bids/bids-mriqc--0.15.1.sing '{inputs}' '{outputs}' participant group",
"dsid": "f367440c-cbcf-11e9-9ad2-002590f97d84",
"exit": 0,
"extra_inputs": [
"containers/images/bids/bids-mriqc--0.15.1.sing"
],
"inputs": [
"sourcedata"
],
"outputs": [
"."
],
"pwd": "."
}
^^^ Do not change lines above ^^^This record could later be reused (by anyone) using datalad rerun to rerun this computation using exactly the same version(s) of input data and the singularity container. You can even now datalad uninstall sourcedata and even containers sub-datasets to save space - they will be retrievable at those exact versions later on if you need to extend or redo your analysis.
- aforementioned example requires DataLad >= 0.11.5 and datalad-containers >= 0.4.0;
- for more eleborate example with use of reproman to parallelize execution on remote resources, see ReproNim/reproman PR#438;
- a copy of the dataset is made available from
///repronim/ds000003-qcand https://github.com/ReproNim/ds000003-qc.
It is a DataLad dataset, so you can either just git clone or datalad install it. You will need to have git-annex available to retrieve any images. And you will need DataLad and datalad-container extension installed for datalad containers-run. Since Singularity is Linux-only application, it will be "functional" only on Linux. On OSX (and possibly Windows), if you have Docker installed, singularity images will be executed through the provided docker shim image.
A few environment variables (in addition to those consulted by datalad and datalad-container) are considered in the scripts of this repository:
The default command (as "hardcoded" in .datalad/config) is run
so running the container executes its default "entry point". Setting
SINGULARITY_CMD=exec makes it possible to run an alternative command
in them (e.g. bash for interactive sessions)::
SINGULARITY_CMD=exec datalad containers-run --explicit -n repronim-reproin bash
and then have datalad record any of the introduced changes. Such
runs will not be reproducible but at least clearly annotated in what
environment corresponding actions were taken.
Development of this project and datalad-container extension was supported by the ReproNim project (NIH 1P41EB019936-01A1). DataLad development was supported by a US-German collaboration in computational neuroscience (CRCNS) "DataGit: converging catalogues, warehouses, and deployment logistics into a federated 'data distribution'" (Halchenko/Hanke), co-funded by the US National Science Foundation (NSF 1429999) and the German Federal Ministry of Education and Research (BMBF 01GQ1411). Additional support is provided by the German federal state of Saxony-Anhalt and the European Regional Development Fund, Project: Center for Behavioral Brain Sciences, Imaging Platform.
All container images are collections of various projects governed by the
corresponding copyrights/licenses. Some are not completely FOSS and might
require additional license(s) to be obtained and provided (e.g. FreeSurfer
license for fmriprep).
Based on the artwork Copyright 2018-2019 Michael Hanke, from myyoda/poster, distributed under CC BY.