From 1b23e7d04d98fcad21ba1836a91c31cd8d921e01 Mon Sep 17 00:00:00 2001
From: Will Graham <32364977+willGraham01@users.noreply.github.com>
Date: Mon, 26 Feb 2024 11:52:57 +0000
Subject: [PATCH] Merging of `bg-atlasapi` and `bg-atlasgen` (#150)
* Write blog post
* bg-atlasapi -> brainglobe-atlasapi
* bg_atlasapi -> brainglobe_atlasapi
* Update atlasgen references
* Link checker
* More link chckers
---
docs/source/about.md | 2 +-
docs/source/blog/atlasapi-atlasgen-merge.md | 42 ++++++
docs/source/community/developers/index.md | 2 +-
.../community/developers/intro_to_codebase.md | 2 +-
.../brainglobe-dependencies.svg | 2 +-
.../repositories/brainglobe-utils/index.md | 2 +-
docs/source/community/external-tools.md | 2 +-
.../adding-a-new-atlas.md | 129 ++++++++----------
.../index.md | 23 ++--
.../usage/atlas-details.md | 0
.../usage/command-line-interface.md | 0
.../usage/python-api.md | 4 +-
.../usage/using-the-files-directly.md | 5 +-
.../documentation/brainglobe-heatmap/index.md | 15 +-
docs/source/documentation/brainreg/index.md | 2 +-
.../documentation/brainreg/installation.md | 2 +-
.../documentation/brainreg/troubleshooting.md | 2 +-
.../source/documentation/brainrender/index.md | 2 +-
.../documentation/brainrender/usage/scene.md | 2 +-
.../usage/using-your-data/registering-data.md | 2 +-
.../documentation/cellfinder/installation.md | 2 +-
docs/source/documentation/index.md | 5 +-
docs/source/index.md | 2 +-
23 files changed, 144 insertions(+), 107 deletions(-)
create mode 100644 docs/source/blog/atlasapi-atlasgen-merge.md
rename docs/source/documentation/{bg-atlasapi => brainglobe-atlasapi}/adding-a-new-atlas.md (60%)
rename docs/source/documentation/{bg-atlasapi => brainglobe-atlasapi}/index.md (85%)
rename docs/source/documentation/{bg-atlasapi => brainglobe-atlasapi}/usage/atlas-details.md (100%)
rename docs/source/documentation/{bg-atlasapi => brainglobe-atlasapi}/usage/command-line-interface.md (100%)
rename docs/source/documentation/{bg-atlasapi => brainglobe-atlasapi}/usage/python-api.md (98%)
rename docs/source/documentation/{bg-atlasapi => brainglobe-atlasapi}/usage/using-the-files-directly.md (78%)
diff --git a/docs/source/about.md b/docs/source/about.md
index d201f500..e6fd8bb1 100644
--- a/docs/source/about.md
+++ b/docs/source/about.md
@@ -23,7 +23,7 @@ We aim to facilitate the access to available datasets, and have developed severa
download gene expression and mesoscale connectomics data for the mouse brain from the Allen institute.
A core step towards facilitating the usage of atlas data was also taken by developing the
-[BrainGlobe AtlasAPI](/documentation/bg-atlasapi/index). This tool provides a
+[BrainGlobe AtlasAPI](/documentation/brainglobe-atlasapi/index). This tool provides a
simple and unified interface for downloading and using data from a number of available atlases. Additionally, new atlases
can easily be added to the API. With this tool we address a major obstacle when developing software for neuroanatomy:
few of the available atlases provide programmatic access to their data, and the APIs used to access the data vary
diff --git a/docs/source/blog/atlasapi-atlasgen-merge.md b/docs/source/blog/atlasapi-atlasgen-merge.md
new file mode 100644
index 00000000..46fc528e
--- /dev/null
+++ b/docs/source/blog/atlasapi-atlasgen-merge.md
@@ -0,0 +1,42 @@
+---
+blogpost: true
+date: Feb 14, 2024
+author: Will Graham
+location: London, England
+category: brainglobe
+language: English
+---
+
+# `bg-atlasapi` and `bg-atlasgen` have merged under a new name
+
+`bg-atlasapi` and `bg-atlasgen` have merged into a single package, now called `brainglobe-atlasapi`.
+`brainglobe-atlasapi` now provides the same API that `bg-atlasapi` provided, in exactly the same way - all that needs to happen is a name change in your scripts.
+
+`bg-atlasgen` used to provide the developer code for adding a new atlas to the collection of BrainGlobe atlases that are available for download.
+This code was exclusively used by developers, however it hinges on the functionality of `bg-atlasapi` to complete the generation and upload process.
+As such, it has now been moved into a submodule, `brainglobe_atlasapi.atlas_generation`, but again the way one interacts with this submodule is the same as they would have with `bg-atlasgen`.
+
+## What do I need to do?
+
+If you are using an install of `bg-atlasapi` that you got through `pip install brainglobe`, then all you need to do is update `brainglobe` to the latest version with
+
+```bash
+pip install --update brainglobe
+```
+
+This will remove the old packages from your environment, install `brainglobe-atlasapi`, and also update all your other BrainGlobe tools to use the new package rather than the old `bg-atlasapi`.
+
+If you were previously using a standalone install of `bg-atlasapi`, to continue receiving updates you should perform the following changes to your environment:
+
+- Uninstall `bg-atlasapi` (and `bg-atlasgen` if you have it).
+- Install the latest version of `brainglobe-atlasapi`.
+- Update all of the following BrainGlobe tools that you also have in your environment:
+ - `brainrender-napari`
+ - `brainglobe-utils`
+ - `brainglobe-segmentation`
+ - `brainreg`
+ - `brainglobe-napari-io`
+ - `cellfinder`
+ - `morphapi`
+ - `brainrender`
+ - `brainglobe-heatmap`
diff --git a/docs/source/community/developers/index.md b/docs/source/community/developers/index.md
index f93ceb65..accf8a5b 100644
--- a/docs/source/community/developers/index.md
+++ b/docs/source/community/developers/index.md
@@ -19,7 +19,7 @@ You can view these repositories and the relevant information by heading to the [
## To contribute a new atlas
-To add a new BrainGlobe atlas, please see the guide [here](/documentation/bg-atlasapi/adding-a-new-atlas).
+To add a new BrainGlobe atlas, please see the guide [here](/documentation/brainglobe-atlasapi/adding-a-new-atlas).
## To contribute code
diff --git a/docs/source/community/developers/intro_to_codebase.md b/docs/source/community/developers/intro_to_codebase.md
index b7ce489e..f1226714 100644
--- a/docs/source/community/developers/intro_to_codebase.md
+++ b/docs/source/community/developers/intro_to_codebase.md
@@ -36,7 +36,7 @@ Code providing functionality related to a specific analysis or visualisation ste
Each BrainGlobe tool has its own Github repository on the BrainGlobe organisation.
Currently stable tools are:
-- [`brainglobe-atlasapi`](https://github.com/brainglobe/bg-atlasapi)
+- [`brainglobe-atlasapi`](https://github.com/brainglobe/brainglobe-atlasapi)
- [`brainglobe-heatmap`](https://github.com/brainglobe/brainglobe-heatmap)
- [`brainglobe-segmentation`](https://github.com/brainglobe/brainglobe-segmentation)
- [`brainreg`](https://github.com/brainglobe/brainreg)
diff --git a/docs/source/community/developers/repositories/brainglobe-meta/brainglobe-dependencies.svg b/docs/source/community/developers/repositories/brainglobe-meta/brainglobe-dependencies.svg
index 5055d444..fcab384a 100644
--- a/docs/source/community/developers/repositories/brainglobe-meta/brainglobe-dependencies.svg
+++ b/docs/source/community/developers/repositories/brainglobe-meta/brainglobe-dependencies.svg
@@ -1,4 +1,4 @@
-
\ No newline at end of file
+
\ No newline at end of file
diff --git a/docs/source/community/developers/repositories/brainglobe-utils/index.md b/docs/source/community/developers/repositories/brainglobe-utils/index.md
index 0f7c410e..e9bafb4f 100644
--- a/docs/source/community/developers/repositories/brainglobe-utils/index.md
+++ b/docs/source/community/developers/repositories/brainglobe-utils/index.md
@@ -29,7 +29,7 @@ The `citation` submodule itself breaks down further to accommodate the steps of
`cite-brainglobe` is only aware of the repositories that we tell it about - and any such repository must have a CITATION.cff (or equivalent metadata file) present in it that we can fetch.
-To make `cite-brainglobe` aware of a new BrainGlobe tool, add a static `Repository` instance to the `citation.repositories` submodule [as detailed here](#citationrepositories), specifying the required information.
+To make `cite-brainglobe` aware of a new BrainGlobe tool, add a static `Repository` instance to the `citation.repositories` submodule as detailed in the `citation.repositories` section, specifying the required information.
### Adding a new supported citation format
diff --git a/docs/source/community/external-tools.md b/docs/source/community/external-tools.md
index b3f1aecf..a9039e23 100644
--- a/docs/source/community/external-tools.md
+++ b/docs/source/community/external-tools.md
@@ -1,6 +1,6 @@
# Software built by the community
-Tools like the [BrainGlobe Atlas API](/documentation/bg-atlasapi/index) were developed to help establish a community of
+Tools like the [BrainGlobe Atlas API](/documentation/brainglobe-atlasapi/index) were developed to help establish a community of
developers creating interoperable tools. The BrainGlobe core developers have created a number of such tools
([cellfinder](/documentation/cellfinder/index), [brainreg](/documentation/brainreg/index),
[brainrender](/documentation/brainrender/index) etc.). More recently, members of the community have built their own
diff --git a/docs/source/documentation/bg-atlasapi/adding-a-new-atlas.md b/docs/source/documentation/brainglobe-atlasapi/adding-a-new-atlas.md
similarity index 60%
rename from docs/source/documentation/bg-atlasapi/adding-a-new-atlas.md
rename to docs/source/documentation/brainglobe-atlasapi/adding-a-new-atlas.md
index d264ff45..28a76d53 100644
--- a/docs/source/documentation/bg-atlasapi/adding-a-new-atlas.md
+++ b/docs/source/documentation/brainglobe-atlasapi/adding-a-new-atlas.md
@@ -2,13 +2,11 @@
## How are BrainGlobe atlases distributed?
-BrainGlobe atlases are downloaded locally to the user’s machine whenever they try to use an atlas that has not yet been
-downloaded.
-By default, they will end up in the `~/.brainglobe` folder,
-unless differently specified in the `bg-atlasapi` configuration.
+BrainGlobe atlases are downloaded locally to the user’s machine whenever they try to use an atlas that has not yet been downloaded.
+By default, they will end up in the `~/.brainglobe` folder, unless differently specified in the `brainglobe-atlasapi` configuration.
The BrainGlobeAtlas class fetches those atlases from a dedicated GIN repository
-([https://gin.g-node.org/brainglobe/atlases](https://gin.g-node.org/brainglobe/atlases)). GIN repos are very similar
+([https://gin.g-node.org/brainglobe/atlases](https://gin.g-node.org/brainglobe/atlases)). GIN repos are very similar
to GitHub repos, but support storage of large datasets, using git-annex.
Only members of the BrainGlobe initiative can upload new atlases to the GIN repo,
and this is essential to ensure the interoperability of all atlases that are part of the BrainGlobe suite.
@@ -16,38 +14,38 @@ Still, we want to keep the process open to external contribution,
and we tried to streamline the process for people who want to expand the list of BrainGlobe supported atlases.
Here’s some information on how the process works, and how you can contribute a new atlas to the initiative:
-The repository of atlases is automatically updated by scripts that reformat atlas data, create a bundled atlas archive
-and upload them. This process will be run every time a new atlas is added or an old atlas is updated. Each atlas is
+The repository of atlases is automatically updated by scripts that reformat atlas data, create a bundled atlas archive
+and upload them. This process will be run every time a new atlas is added or an old atlas is updated. Each atlas is
generated in a separate script, so if you want to contribute with a new atlas:
-* Fork [github.com/brainglobe/bg-atlasgen](https://github.com/brainglobe/bg-atlasgen)
-* Create an atlas generation script in the `atlas_scripts` directory (see below)
+* Fork [github.com/brainglobe/brainglobe-atlasapi](https://github.com/brainglobe/brainglobe-atlasapi)
+* Create an atlas generation script in the `atlas_generation/atlas_scripts` directory (see below)
* Submit a pull request back to the original repository.
-If you are new to GitHub, or you have any questions about writing your script, feel free to reach out via a
-[GitHub issue](https://github.com/brainglobe/bg-atlasgen/issues) or by starting a discussion using the
-[BrainGlobe tag over at the image.sc forum](https://forum.image.sc/tag/brainglobe) and we’ll be very happy to provide
+If you are new to GitHub, or you have any questions about writing your script, feel free to reach out via a
+[GitHub issue](https://github.com/brainglobe/brainglobe-atlasapi/issues) or by starting a discussion using the
+[BrainGlobe tag over at the image.sc forum](https://forum.image.sc/tag/brainglobe) and we’ll be very happy to provide
you with all the assistance you need to contribute to the project!
## What data do you need to contribute an atlas to the BrainGlobe Initiative?
-To contribute your favourite atlas in a BrainGlobe-compatible format, these are the fundamental ingredients you’ll
+To contribute your favourite atlas in a BrainGlobe-compatible format, these are the fundamental ingredients you’ll
need to have before starting:
* Information to credit the atlas: a publication, if available, and a website of the atlas, if available.
-* A reference 3D stack in your favorite format, as long as you can load it to a numpy array. This will be the reference
-stack, used to represent the anatomy of the brain. If you have more than one reference brain (e.g., multiple transgenic
-lines that people can use for registrations), don’t worry: just choose the most important one, and the others can be
+* A reference 3D stack in your favorite format, as long as you can load it to a numpy array. This will be the reference
+stack, used to represent the anatomy of the brain. If you have more than one reference brain (e.g., multiple transgenic
+lines that people can use for registrations), don’t worry: just choose the most important one, and the others can be
distributed via BrainGlobe as well.
-* An annotation 3D stack that contains 0s in all non-annotated voxels, and integer numbers that correspond to the ids
-of all the different regions. IDs should refer to the finest level of your region hierarchy. For example, in a brain
-where the telecephalon has ID 1, the cortex ID 2, and the visual cortex ID 3, a voxel in the visual cortex should just
-have 3. The BrainGlobe Atlas API will know how to take care of pooling all voxels that belong to the cortex using the
+* An annotation 3D stack that contains 0s in all non-annotated voxels, and integer numbers that correspond to the ids
+of all the different regions. IDs should refer to the finest level of your region hierarchy. For example, in a brain
+where the telecephalon has ID 1, the cortex ID 2, and the visual cortex ID 3, a voxel in the visual cortex should just
+have 3. The BrainGlobe Atlas API will know how to take care of pooling all voxels that belong to the cortex using the
annotation and the region hierarchy information (more on that later).
-* A description of all the structures of the brain and their hierarchy. This represented in BrainGlobe with a list of
+* A description of all the structures of the brain and their hierarchy. This represented in BrainGlobe with a list of
dictionaries where each dictionary describes a region with the following keys:
-```
+```python
STRUCTURE_TEMPLATE = {
"acronym": "VIS", # shortened name of the region
"id": 3, # region id
@@ -57,12 +55,12 @@ STRUCTURE_TEMPLATE = {
}
```
-The `structure_id_path` is the fundamental key argument for bg-atlasapi to understand the hierarchy of your regions.
-Going back to our example above, if telencephalon is 1, cortex 2, and visual cortex 3, `structure_id_path` for the
-cortex region will be \[1, 2], and for the visual cortex region will be \[1, 2, 3]. you don’t necessarily need a fixed
-number of levels in your hierarchy. You should just make sure that there is one and only one root region (i.e., a region
-with `structure_id_path` of length 1). If your atlas does not have a multilevel hierarchy, just create a root region
-that all other regions are children of. How to generate the region hierarchy is up to you; just make sure you have a
+The `structure_id_path` is the fundamental key argument for `brainglobe-atlasapi` to understand the hierarchy of your regions.
+Going back to our example above, if telencephalon is 1, cortex 2, and visual cortex 3, `structure_id_path` for the
+cortex region will be \[1, 2], and for the visual cortex region will be \[1, 2, 3]. you don’t necessarily need a fixed
+number of levels in your hierarchy. You should just make sure that there is one and only one root region (i.e., a region
+with `structure_id_path` of length 1). If your atlas does not have a multilevel hierarchy, just create a root region
+that all other regions are children of. How to generate the region hierarchy is up to you; just make sure you have a
way of generating this list of dictionaries in python when you’ll start creating the atlas.
:::{caution}
@@ -74,7 +72,7 @@ Do not use 0 (zero) as the id of a structure, because the annotation stack will
**Atlas generation takes place in two steps:**
1. Generate an “Atlas script” written in python
-2. Use the atlas generation code [available from BrainGlobe](https://github.com/brainglobe/bg-atlasgen/tree/main/bg\_atlasgen)
+2. Use the atlas generation code [available from BrainGlobe](https://github.com/brainglobe/brainglobe-atlasapi/tree/main/brainglobe_atlasapi/atlas_generation/)
3. to generate the atlas from the atlas script.
### Atlas scripts
@@ -85,7 +83,6 @@ Do not use 0 (zero) as the id of a structure, because the annotation stack will
2. To load the atlas data (e.g. reference image).
-
**The metadata that needs to be specified in the atlas script is:**
* RES\_UM: resolution of the atlas in micrometers (voxel spacing)
@@ -98,86 +95,74 @@ Do not use 0 (zero) as the id of a structure, because the annotation stack will
* \_\_version\_\_: the version number of the atlas: when atlases get updated/modified new versions are released.
* root structure id: ID of the root (base) brain region in the hierarchy of brain regions.
-This metadata is used to provide information about the atlas (e.g., which species is it about), to give credit to the
+This metadata is used to provide information about the atlas (e.g., which species is it about), to give credit to the
original creators of the atlas data and to provide data used by other BrainGlobe tools (e.g., the atlas orientation).
-
**In addition to laying out the metadata, atlas scripts are being used to load (or download) the actual atlas data:**
* Template image volume: a 3D image with the atlas’ template
* Annotated image volume: a 3D image where each voxel is annotated with the ID of the atlas brain region at that point
* hemispheres stack (optional): a 3D image where each voxel is annotated with the ID of the hemisphere it belongs to
* Structures list: a list of brain regions (structures) names
-* Meshes dict: a dictionary with entries of the type structure->file path specifying the path to the .obj mesh file
+* Meshes dict: a dictionary with entries of the type structure->file path specifying the path to the .obj mesh file
of each brain region
### Atlas generation
-Once an atlas script has been written which sets out all the necessary metadata and loads all the files required,
-code provided by BrainGlobe’s atlas generation repository can be used to format the data as needed by the BrainGlobe
-AtlasAPI. Atlasgen provides one convenient function `wrapup_atlas_from_data` which takes all the metadata and data
-from above and creates the data structures required. These data structures include .tiff files with the various images,
-csv files with the structures metadata information, a README file laying out the atlas content, etc. The same function
+
+Once an atlas script has been written which sets out all the necessary metadata and loads all the files required,
+code provided by BrainGlobe’s atlas generation repository can be used to format the data as needed by the BrainGlobe
+AtlasAPI. Atlasgen provides one convenient function `wrapup_atlas_from_data` which takes all the metadata and data
+from above and creates the data structures required. These data structures include .tiff files with the various images,
+csv files with the structures metadata information, a README file laying out the atlas content, etc. The same function
creates a compressed folder which can be loaded to GIN for distribution (see above).
### Extracting meshes
-While most atlases will have a set of 3D images that can be used as annotated and template stacks,
-not all atlases will have mesh (.obj) files for each brain region in the atlas.
-However, regions’ meshes are used frequently by BrainGlobe tools,
-especially in `brainrender` for visualizing anatomical data in 3D.
-For this reason,
-the atlas generation code from BrainGlobe provides functionality that can be used to generate such mesh files.
-A mask image is created which only contains the annotation for the brain region of interest,
-and a marching cubes algorithm is used to identify the surface of the region;
- finally, a mesh is created from the output of the marching cubes algorithm, and the results are saved to a .obj file.
+
+While most atlases will have a set of 3D images that can be used as annotated and template stacks, not all atlases will have mesh (.obj) files for each brain region in the atlas.
+However, regions’ meshes are used frequently by BrainGlobe tools, especially in `brainrender` for visualizing anatomical data in 3D.
+For this reason, the atlas generation code from BrainGlobe provides functionality that can be used to generate such mesh files.
+A mask image is created which only contains the annotation for the brain region of interest, and a marching cubes algorithm is used to identify the surface of the region; finally, a mesh is created from the output of the marching cubes algorithm, and the results are saved to a .obj file.
Repeating the process for each region in the atlas, it is possible to easily create all the necessary meshes.
### How can I be sure I get credited for the atlas I contributed?
-Included with the atlas metadata, which is specified during atlas creation (see above), is a link to any online
-resource describing the atlas and to a publication associated with the atlas.
-This ensures that information about the original creators of the atlas data accompanies the atlas data at all time,
-so that credit can easily be given to the atlas creators.
+Included with the atlas metadata, which is specified during atlas creation (see above), is a link to any online resource describing the atlas and to a publication associated with the atlas.
+
+This ensures that information about the original creators of the atlas data accompanies the atlas data at all time, so that credit can easily be given to the atlas creators.
-Sometimes, the atlas creator and the person contributing the atlas to BrainGlobe (i.e., going through the atlas
-generation process laid out here) may not be the same. The `ATLAS_PACKAGER` argument in the metadata allow
-specifying the name and contact of the person who curated the BrainGlobe atlas creation.
+Sometimes, the atlas creator and the person contributing the atlas to BrainGlobe (i.e., going through the atlas generation process laid out here) may not be the same.
+The `ATLAS_PACKAGER` argument in the metadata allow specifying the name and contact of the person who curated the BrainGlobe atlas creation.
### Inspecting results
+
To check if everything went smoothly,
you can just go to the folder with the atlas content
(named `atlasname_resolutionum` in the directory you were generating the atlas in) and check out its content.
For the metadata, you can just open the json file and inspect it.
-To inspect the reference.tiff file and the annotation.tiff file, you can just drag and drop them in napari
-(to install napari, see [here](https://napari.org/stable/tutorials/fundamentals/installation)),
-and they will open as an image layer, and a label layer, respectively. You know the orientation is correct if you are
-looking at upright (dorsal top, ventral bottom) frontal sections, and when you scroll the slider to inspect sections
-with a higher index, you are moving from anterior to posterior.
+To inspect the reference.tiff file and the annotation.tiff file, you can just drag and drop them in napari (to install napari, see [here](https://napari.org/stable/tutorials/fundamentals/installation)), and they will open as an image layer, and a label layer, respectively.
+You know the orientation is correct if you are looking at upright (dorsal top, ventral bottom) frontal sections, and when you scroll the slider to inspect sections with a higher index, you are moving from anterior to posterior.
To inspect the meshes, BrainGlobe provides a simple tool
load and visualize a set of .obj files for quick inspection, e.g.:
```python
-from bg_atlasgen.mesh_utils import inspect_meshes_folder
+from brainglobe_atlasapi.atlas_generation.mesh_utils import inspect_meshes_folder
inspect_meshes_folder("~/.brainglobe/temp/allen_mouse_10um_v1.0/meshes")
```
-
-Once an atlas is created with BrainGlobe’s atlas generation tools,
-it can be used with most software from the BrainGlobe software suite, including [brainrender](/documentation/brainrender/index)
-which provides a convenient GUI for visually inspecting the generated atlas meshes.
+Once an atlas is created with BrainGlobe’s atlas generation tools, it can be used with most software from the BrainGlobe software suite.
+This includes [brainrender](/documentation/brainrender/index) which provides a convenient GUI for visually inspecting the generated atlas meshes.
## Uploading the atlas
-After the pull request is merged, one of the BrainGlobe core developers will run the script and upload the packaged
-atlas (`.tar.gz` archive) to [the GIN repository](https://gin.g-node.org/BrainGlobe/atlases). The
-[`latest_versions.conf`](https://gin.g-node.org/BrainGlobe/atlases/src/master/last_versions.conf) file will also be
-updated to include the new atlas. At this point, the atlas will be available for use in the API.
+After the pull request is merged, one of the BrainGlobe core developers will run the script and upload the packaged atlas (`.tar.gz` archive) to [the GIN repository](https://gin.g-node.org/BrainGlobe/atlases).
+The [`latest_versions.conf`](https://gin.g-node.org/BrainGlobe/atlases/src/master/last_versions.conf) file will also be updated to include the new atlas. At this point, the atlas will be available for use in the API.
## Updating an existing atlas
-To update an existing atlas, a pull request must be raised to change the existing script. Once this pull request is
-merged, the new atlas will be generated and uploaded to the GIN repository as normal. The
-[`latest_versions.conf`](https://gin.g-node.org/BrainGlobe/atlases/src/master/last_versions.conf) can then be updated
-to reflect the latest version.
\ No newline at end of file
+
+To update an existing atlas, a pull request must be raised to change the existing script.
+Once this pull request is merged, the new atlas will be generated and uploaded to the GIN repository as normal.
+The [`latest_versions.conf`](https://gin.g-node.org/BrainGlobe/atlases/src/master/last_versions.conf) can then be updated to reflect the latest version.
diff --git a/docs/source/documentation/bg-atlasapi/index.md b/docs/source/documentation/brainglobe-atlasapi/index.md
similarity index 85%
rename from docs/source/documentation/bg-atlasapi/index.md
rename to docs/source/documentation/brainglobe-atlasapi/index.md
index 4a868c9d..c7ef47b3 100644
--- a/docs/source/documentation/bg-atlasapi/index.md
+++ b/docs/source/documentation/brainglobe-atlasapi/index.md
@@ -1,10 +1,11 @@
-# BrainGlobe Atlas API (bg-atlasapi)
-Many excellent brain atlases exist for different species. Some of them have an API (application programming interface)
-to allow users to interact with the data programmatically (e.g. the excellent
-[Allen Mouse Brain Atlas](https://portal.brain-map.org)), but many do not, and there is no consistent way to process
+# BrainGlobe Atlas API (brainglobe-atlasapi)
+
+Many excellent brain atlases exist for different species. Some of them have an API (application programming interface)
+to allow users to interact with the data programmatically (e.g. the excellent
+[Allen Mouse Brain Atlas](https://portal.brain-map.org)), but many do not, and there is no consistent way to process
data from multiple sources.
-The brainglobe atlas API (BG-AtlasAPI) deals with this problem by providing a common interface for programmers to download and process data from multiple sources.
+The brainglobe atlas API deals with this problem by providing a common interface for programmers to download and process data from multiple sources.
Each atlas consists of data files in a common format:
@@ -33,14 +34,14 @@ A number of atlases are in development, but those available currently are:
## Installation
-BG-AtlasAPI works with Python >3.6, and can be installed from PyPI with:
+BrainGlobe AtlasAPI works with Python >3.6, and can be installed from PyPI with:
```bash
-pip install bg-atlasapi
+pip install brainglobe-atlasapi
```
-
## Usage
+
```{toctree}
:maxdepth: 2
usage/atlas-details
@@ -51,14 +52,16 @@ usage/using-the-files-directly
```
## More details
+
```{toctree}
:maxdepth: 1
adding-a-new-atlas
```
## Citation
+
If you find the BrainGlobe Atlas API useful, please cite the paper in your work:
->Claudi, F., Petrucco, L., Tyson, A. L., Branco, T., Margrie, T. W. and Portugues, R. (2020). BrainGlobe Atlas API: a common interface for neuroanatomical atlases. Journal of Open Source Software, 5(54), 2668, https://doi.org/10.21105/joss.02668
+>Claudi, F., Petrucco, L., Tyson, A. L., Branco, T., Margrie, T. W. and Portugues, R. (2020). BrainGlobe Atlas API: a common interface for neuroanatomical atlases. Journal of Open Source Software, 5(54), 2668,
-**Don't forget to cite the developers of the atlas that you used!**
\ No newline at end of file
+**Don't forget to cite the developers of the atlas that you used!**
diff --git a/docs/source/documentation/bg-atlasapi/usage/atlas-details.md b/docs/source/documentation/brainglobe-atlasapi/usage/atlas-details.md
similarity index 100%
rename from docs/source/documentation/bg-atlasapi/usage/atlas-details.md
rename to docs/source/documentation/brainglobe-atlasapi/usage/atlas-details.md
diff --git a/docs/source/documentation/bg-atlasapi/usage/command-line-interface.md b/docs/source/documentation/brainglobe-atlasapi/usage/command-line-interface.md
similarity index 100%
rename from docs/source/documentation/bg-atlasapi/usage/command-line-interface.md
rename to docs/source/documentation/brainglobe-atlasapi/usage/command-line-interface.md
diff --git a/docs/source/documentation/bg-atlasapi/usage/python-api.md b/docs/source/documentation/brainglobe-atlasapi/usage/python-api.md
similarity index 98%
rename from docs/source/documentation/bg-atlasapi/usage/python-api.md
rename to docs/source/documentation/brainglobe-atlasapi/usage/python-api.md
index 3e737f67..a5f07700 100644
--- a/docs/source/documentation/bg-atlasapi/usage/python-api.md
+++ b/docs/source/documentation/brainglobe-atlasapi/usage/python-api.md
@@ -7,7 +7,7 @@ version of this atlas files will be downloaded from the [remote GIN repository](
and stored on your local machine (by default, in `~/.brainglobe`):
```python
-from bg_atlasapi import BrainGlobeAtlas
+from brainglobe_atlasapi import BrainGlobeAtlas
from pprint import pprint
bg_atlas = BrainGlobeAtlas("allen_mouse_100um", check_latest=False)
@@ -17,7 +17,7 @@ To know what atlases are available through BrainGlobe, we can use the `show_atla
(requires an internet connection):
```python
-from bg_atlasapi import show_atlases
+from brainglobe_atlasapi import show_atlases
show_atlases()
```
diff --git a/docs/source/documentation/bg-atlasapi/usage/using-the-files-directly.md b/docs/source/documentation/brainglobe-atlasapi/usage/using-the-files-directly.md
similarity index 78%
rename from docs/source/documentation/bg-atlasapi/usage/using-the-files-directly.md
rename to docs/source/documentation/brainglobe-atlasapi/usage/using-the-files-directly.md
index c659d1ed..c4894fea 100644
--- a/docs/source/documentation/bg-atlasapi/usage/using-the-files-directly.md
+++ b/docs/source/documentation/brainglobe-atlasapi/usage/using-the-files-directly.md
@@ -1,8 +1,8 @@
# Using the files directly
-Although the API should provide you with the tools you need to work with the atlas \(and if they don't, please [raise an issue](https://github.com/brainglobe/bg-atlasapi/issues)\), you may wish to find the files themselves.
+Although the API should provide you with the tools you need to work with the atlas (and if they don't, please [raise an issue](https://github.com/brainglobe/brainglobe-atlasapi/issues)), you may wish to find the files themselves.
-By default, atlases will be downloaded and saved into your home directory in a hidden directory \(`.brainglobe`\), with one directory per atlas e.g. `~/.brainglobe/allen_mouse_10um_v0.3`\).
+By default, atlases will be downloaded and saved into your home directory in a hidden directory (`~/.brainglobe`), with one directory per atlas e.g. `~/.brainglobe/allen_mouse_10um_v0.3`.
In each atlas directory, there will be the following subdirectories and files:
@@ -12,4 +12,3 @@ In each atlas directory, there will be the following subdirectories and files:
* `structures.json` A file describing the mapping of brain region ID to region name, and the hierarchy of brain structures
* `metadata.json` A file containing the atlas metadata, such as the shape of the images, the anatomical orientation, and the details of the source of the atlas
* `README.txt` A human-readable version of the metadata and brain region hierarchies
-
diff --git a/docs/source/documentation/brainglobe-heatmap/index.md b/docs/source/documentation/brainglobe-heatmap/index.md
index f4ebf82b..86a263b7 100644
--- a/docs/source/documentation/brainglobe-heatmap/index.md
+++ b/docs/source/documentation/brainglobe-heatmap/index.md
@@ -1,4 +1,5 @@
# brainglobe-heatmap
+
`brainglobe-heatmap` allows you to create heatmaps, mapping scalar values for each brain region (e.g., number of labelled cells in each region) to a color and creating beautiful visualizations in 2D (using [matplotlib](https://matplotlib.org/) or 3D (using [brainrender](https://brainglobe.info/documentation/brainrender/index.html)).
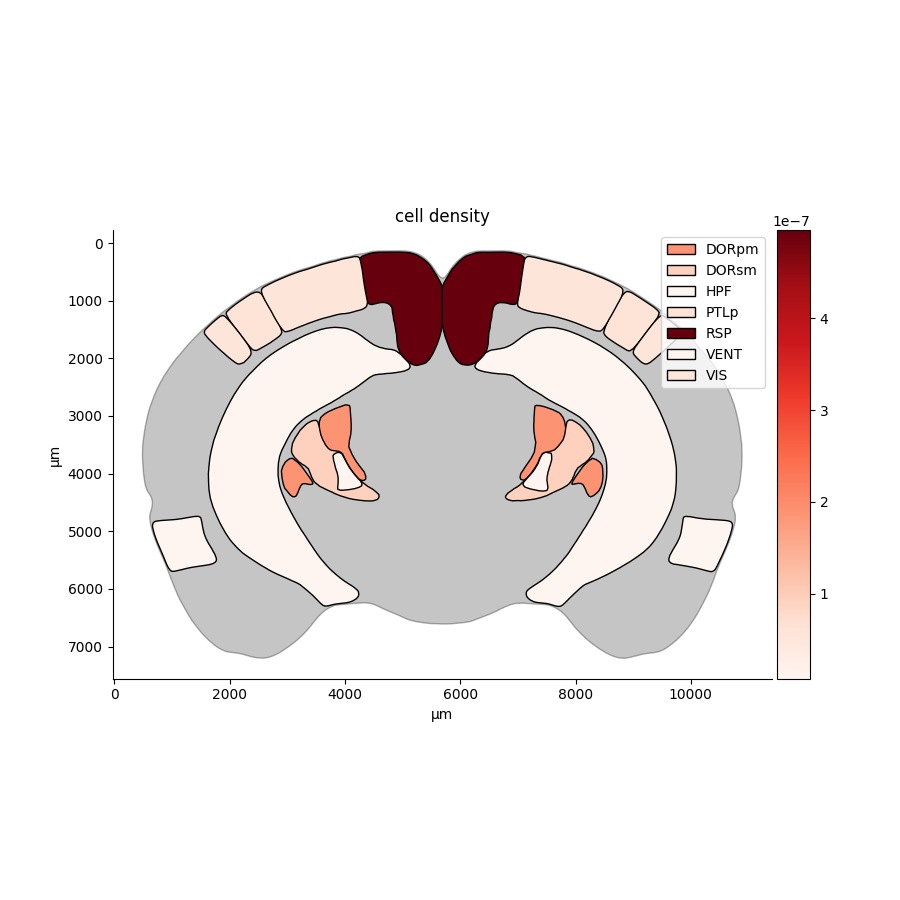
@@ -10,10 +11,11 @@
**3D heatmap generated using brainrender**
## Installation
-`pip install brainglobe-heatmap`
+`pip install brainglobe-heatmap`
## User guide
+
The starting point for a heatmap visualization is a `dict` assigning scalar values to a set of brain regions (identified by their acronym).
For example:
@@ -47,6 +49,7 @@ To keep a section of the 3D brain, two planes with normal vectors facing in oppo
and everything in-between the two planes is kept as a slice.
### Slicing plane position
+
Finding the right position and orientation to the plane can take some tweaking. `brainglobe-heatmap` provides a `planner` class that makes the process easier by showing the position of the planes and how they intersect with the user provided regions (see image above).
In `examples/plan.py` there's an example showing how to use the `planner`:
@@ -83,6 +86,7 @@ f = bgh.heatmap(
```
Also, you can create a slice with a plane centered in the brain by passing `position=None`:
+
```python
f = bgh.heatmap(
values,
@@ -96,6 +100,7 @@ f = bgh.heatmap(
```
### Visualization
+
Once happy with the position of the slicing planes, creating a visualization is as simple as:
```python
@@ -119,8 +124,8 @@ bgh.heatmap(
Here, `format` specifies if a 2D plot should be made (using `matplotlib`) or a 3D rendering instead (using `brainrender`). The `cmap` parameter specifies the colormap used and `vmin, vmax` the color range.
### Regions coordinates
-You can use `brainglobe-heatmap` to get the coordinates of the 2D 'slices' (in the 2D plane's coordinates system):
+You can use `brainglobe-heatmap` to get the coordinates of the 2D 'slices' (in the 2D plane's coordinates system):
```python
@@ -138,15 +143,17 @@ coordinates = bgh.get_plane_coordinates(
)
```
-## Using `brainglobe-heatmap` with other atlases.
+## Using `brainglobe-heatmap` with other atlases
`brainglobe-heatmap` uses `brainrender` which, in turn, uses brainglobe's `Atlas API` under the hood. That means that all of `brainglobe-heatmap`'s functionality is compatible with any of the atlases supported by the atlas API. `bgh.heatmap`, `bgh.planner` and `bgh.get_plane_coordinates` all accept a `atlas_name` argument, pass the name of the atlas name you'd like to use!
-For more information see the API's [documentation](https://brainglobe.info/documentation/bg-atlasapi/index.html).
+For more information see the API's [documentation](../brainglobe-atlasapi/index.md).
## Contributing
+
Contributions to `brainglobe-heatmap` are more than welcome. Please see the [Developer's guide](https://github.com/brainglobe/.github/blob/main/CONTRIBUTING.md).
## Citing `brainglobe-heatmap`
+
If you use `brainglobe-heatmap` in your work, please cite it as:
```
diff --git a/docs/source/documentation/brainreg/index.md b/docs/source/documentation/brainreg/index.md
index 5e1807c7..2f80116c 100644
--- a/docs/source/documentation/brainreg/index.md
+++ b/docs/source/documentation/brainreg/index.md
@@ -2,7 +2,7 @@
brainreg is an update to [amap](https://github.com/SainsburyWellcomeCentre/amap_python) (itself a port of the
[original Java software](https://www.nature.com/articles/ncomms11879)) to include multiple registration backends,
-and to support the many atlases provided by [bg-atlasapi](https://github.com/brainglobe/bg-atlasapi).
+and to support the many atlases provided by [brainglobe-atlasapi](https://github.com/brainglobe/brainglobe-atlasapi).
Currently, the only registration backend is [NiftyReg](http://cmictig.cs.ucl.ac.uk/wiki/index.php/NiftyReg).
diff --git a/docs/source/documentation/brainreg/installation.md b/docs/source/documentation/brainreg/installation.md
index d9aa4dc6..3f204013 100644
--- a/docs/source/documentation/brainreg/installation.md
+++ b/docs/source/documentation/brainreg/installation.md
@@ -76,4 +76,4 @@ If you are using macOS then please also run `conda install -c conda-forge niftyr
When brainreg runs, the appropriate reference atlas will be downloaded (if not previously
done so). To install download BrainGlobe atlases in advance, please see the guide to
-[the BrainGlobe Atlas API command-line interface](/documentation/bg-atlasapi/usage/command-line-interface) for details.
\ No newline at end of file
+[the BrainGlobe Atlas API command-line interface](/documentation/brainglobe-atlasapi/usage/command-line-interface) for details.
\ No newline at end of file
diff --git a/docs/source/documentation/brainreg/troubleshooting.md b/docs/source/documentation/brainreg/troubleshooting.md
index 1552d723..72b77801 100644
--- a/docs/source/documentation/brainreg/troubleshooting.md
+++ b/docs/source/documentation/brainreg/troubleshooting.md
@@ -12,7 +12,7 @@ orientation and the voxel sizes](/documentation/setting-up/image-definition). Pl
## Improving registration performance
There are many ways to improve registration performance, but this will depend on your data. Some options to try include:
* [Changing registration parameters](user-guide/parameters)
-* Registration to a [different resolution atlas](/documentation/bg-atlasapi/usage/atlas-details) (if available)
+* Registration to a [different resolution atlas](/documentation/brainglobe-atlasapi/usage/atlas-details) (if available)
* Improving data quality. brainreg relies on high-contrast data. Poorly cleared tissue, or tissue with limited
autofluorescence may not register well
diff --git a/docs/source/documentation/brainrender/index.md b/docs/source/documentation/brainrender/index.md
index d7bec7ea..964f9e48 100644
--- a/docs/source/documentation/brainrender/index.md
+++ b/docs/source/documentation/brainrender/index.md
@@ -8,7 +8,7 @@
A core design goal is to facilitate the rendering of any data registered to a reference atlas. To this end,
`brainrender` facilitates the creation of 3D objects from many different types of data (e.g. cell locations,
brain regions) within minimal need for the development of dedicated code. In addition, `brainrender` is fully
-integrated with the [BrainGlobe Atlas API](/documentation/bg-atlasapi/index) ensuring that you can use `brainrender`
+integrated with the [BrainGlobe Atlas API](/documentation/brainglobe-atlasapi/index) ensuring that you can use `brainrender`
with any atlas supported by the API with no need for any changes in your code.

diff --git a/docs/source/documentation/brainrender/usage/scene.md b/docs/source/documentation/brainrender/usage/scene.md
index 81461b48..0628a66a 100644
--- a/docs/source/documentation/brainrender/usage/scene.md
+++ b/docs/source/documentation/brainrender/usage/scene.md
@@ -5,7 +5,7 @@ visualization, to add brain regions meshes (by interacting with the `Atlas` clas
it to export images (using the `Render` class). Finally, the creation of a `Scene` is the first step towards the
generation of [videos and animations](videos-animations-and-exporting-to-html) as well.
-Given its integration with the [BrainGlobe Atlas API](/documentation/bg-atlasapi/index), `brainrender` can be used with
+Given its integration with the [BrainGlobe Atlas API](/documentation/brainglobe-atlasapi/index), `brainrender` can be used with
any atlas supported by the API. The moment when you're creating your `Scene` is when you have a chance to specify
which atlas you intend to use by passing `atlas_name='atlas to use'` to `Scene()`.
diff --git a/docs/source/documentation/brainrender/usage/using-your-data/registering-data.md b/docs/source/documentation/brainrender/usage/using-your-data/registering-data.md
index 8ca87b6d..cccb49e1 100644
--- a/docs/source/documentation/brainrender/usage/using-your-data/registering-data.md
+++ b/docs/source/documentation/brainrender/usage/using-your-data/registering-data.md
@@ -1,7 +1,7 @@
# Registering data
To get the most out of brainrender, your data should be registered to one of the atlases supported by
-the [BrainGlobe Atlas API](/documentation/bg-atlasapi/index). If you've used BrainGlobe's software for processing your
+the [BrainGlobe Atlas API](/documentation/brainglobe-atlasapi/index). If you've used BrainGlobe's software for processing your
raw data (e.g. [brainreg](/documentation/brainreg/index)) your data will already be registered, and you need not
worry about any of this. If not, then some steps are necessary for registering your data.
diff --git a/docs/source/documentation/cellfinder/installation.md b/docs/source/documentation/cellfinder/installation.md
index 33666d9c..00466f90 100644
--- a/docs/source/documentation/cellfinder/installation.md
+++ b/docs/source/documentation/cellfinder/installation.md
@@ -23,7 +23,7 @@ pip install cellfinder[napari]>=1.0.0 # Run this if you want to install the API
### Installing BrainGlobe Atlases
-To install download BrainGlobe atlases in advance, please see the guide to [the BrainGlobe Atlas API command-line interface](/documentation/bg-atlasapi/usage/command-line-interface).
+To install download BrainGlobe atlases in advance, please see the guide to [the BrainGlobe Atlas API command-line interface](/documentation/brainglobe-atlasapi/usage/command-line-interface).
### Installing `brainmapper`
diff --git a/docs/source/documentation/index.md b/docs/source/documentation/index.md
index ad96508d..0b257520 100644
--- a/docs/source/documentation/index.md
+++ b/docs/source/documentation/index.md
@@ -8,6 +8,7 @@ Your environment should run Python 3.9 or 3.10. To specify the Python version fo
```bash
conda create -n brainglobe-env python=3.10
```
+
Once you have created and activated your desired environment, you can install all BrainGlobe tools using `pip`:
```bash
@@ -55,9 +56,9 @@ Once you have installed `brainglobe`, or [installed an individual tool](#install
```{toctree}
:maxdepth: 1
setting-up/index
-brainglobe-utils/citation-module
-bg-atlasapi/index
+brainglobe-atlasapi/index
brainglobe-space/index
+brainglobe-utils/citation-module
brainreg/index
brainglobe-segmentation/index
brainglobe-workflows/index
diff --git a/docs/source/index.md b/docs/source/index.md
index cd2f6a01..8a23414a 100644
--- a/docs/source/index.md
+++ b/docs/source/index.md
@@ -7,7 +7,7 @@ We have three aims:
* Develop specialist software for specific analysis and visualisation needs, such as
[cellfinder](/documentation/cellfinder/index) and [brainrender](/documentation/brainrender/index).
* Develop core tools to facilitate [others to build interoperable tools in Python](community/external-tools.md), e.g., the
-[BrainGlobe Atlas API](/documentation/bg-atlasapi/index).
+[BrainGlobe Atlas API](/documentation/brainglobe-atlasapi/index).
* Build a community of neuroscientists and developers to share knowledge, build software and engage with the
scientific, and open-source community (e.g., by organising hackathons).