How do I interpret tutorial heatmap-dotplot? #496
-
|
Hi all, The updated tutorial of SCENIC+ goes over creating a heatmap-dotplot in the downstream analysis of the scplus_mudata output, where "the color represent target gene enrichment and the dotsize represent target region enrichment" (shown below). I have a difficult time understanding what "enrichment" exactly means. Does it just mean higher expression of the target gene and higher target region accessibility? If so, if we see a high enrichment of both in a cell type for a -/- repressor eRegulon, does it mean that eRegulon is downregulated in that cell type? (Eg. TCF12 -/- in BG cell type). I'm struggling with how I should select eRegulons that may be an important regulator in a specific cell type with this kind of heatmap-dotplot. Best, |
Beta Was this translation helpful? Give feedback.
Replies: 1 comment
-
|
Just passing by, from my perspective it refers to the AUC algorithm, which is rank-based and used to estimate the relative expression or accessibility of the gene or region sets. |
Beta Was this translation helpful? Give feedback.
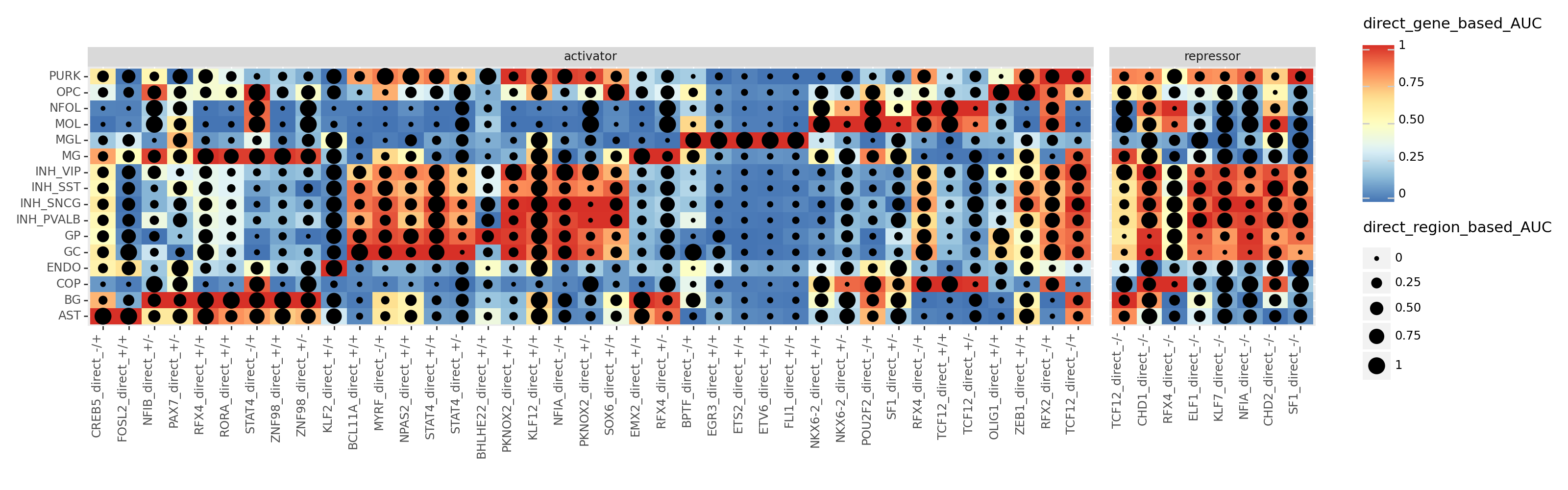
Just passing by, from my perspective it refers to the AUC algorithm, which is rank-based and used to estimate the relative expression or accessibility of the gene or region sets.