A GUI-based Python framework for segmentation, tracking, cell cycle annotations and quantification of microscopy data
Written in Python 3 by Francesco Padovani and Benedikt Mairhoermann.
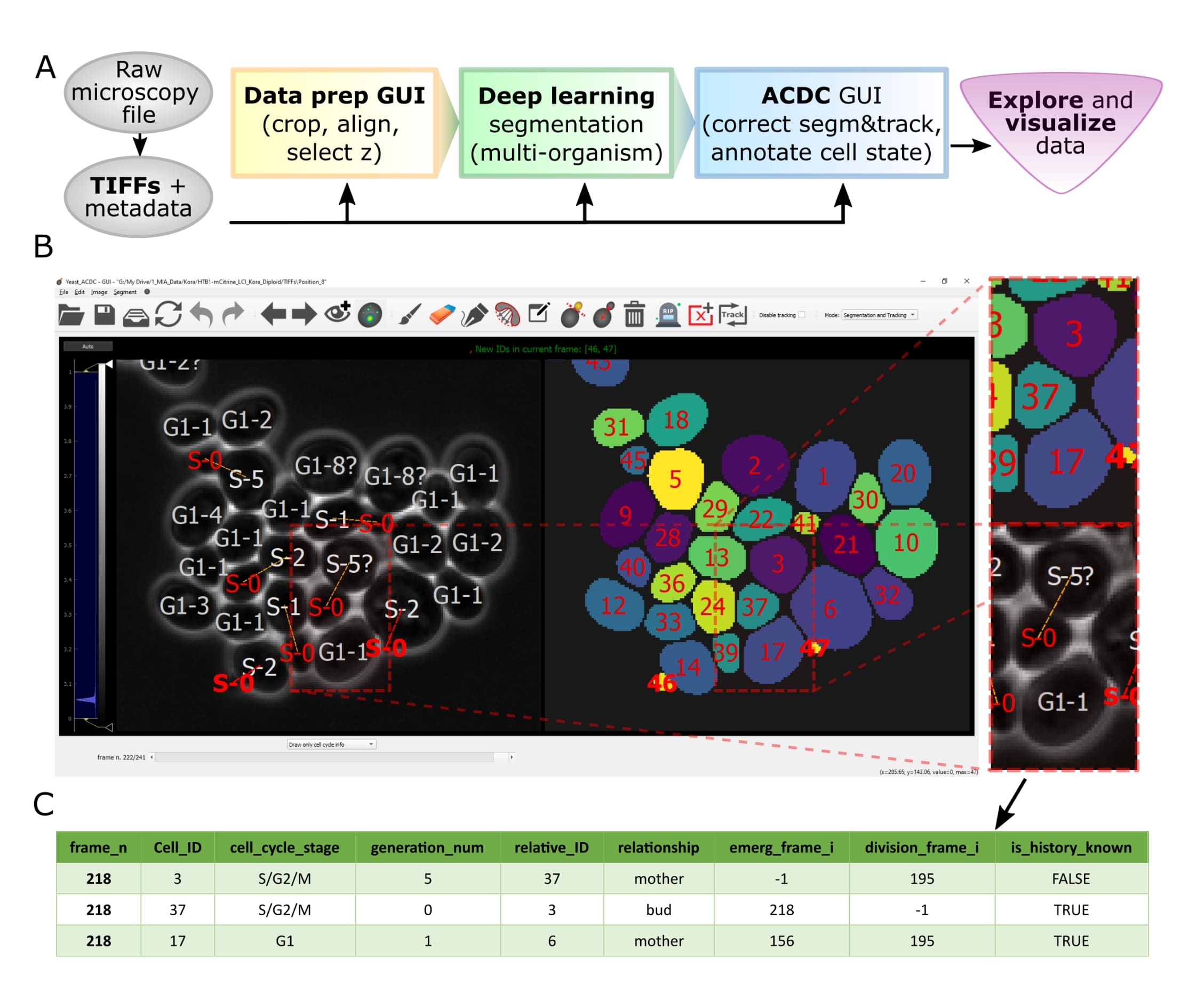
Overview of pipeline and GUI
Let's face it, when dealing with segmentation of microscopy data we often do not have time to check that everything is correct, because it is a tedious and very time consuming process. Cell-ACDC comes to the rescue! We combined the currently best available neural network models (such as Segment Anything Model (SAM), YeaZ, cellpose, StarDist, YeastMate, omnipose, delta, DeepSea, etc.) and we complemented them with a fast and intuitive GUI.
We developed and implemented several smart functionalities such as real-time continuous tracking, automatic propagation of error correction, and several tools to facilitate manual correction, from simple yet useful brush and eraser to more complex flood fill (magic wand) and Random Walker segmentation routines.
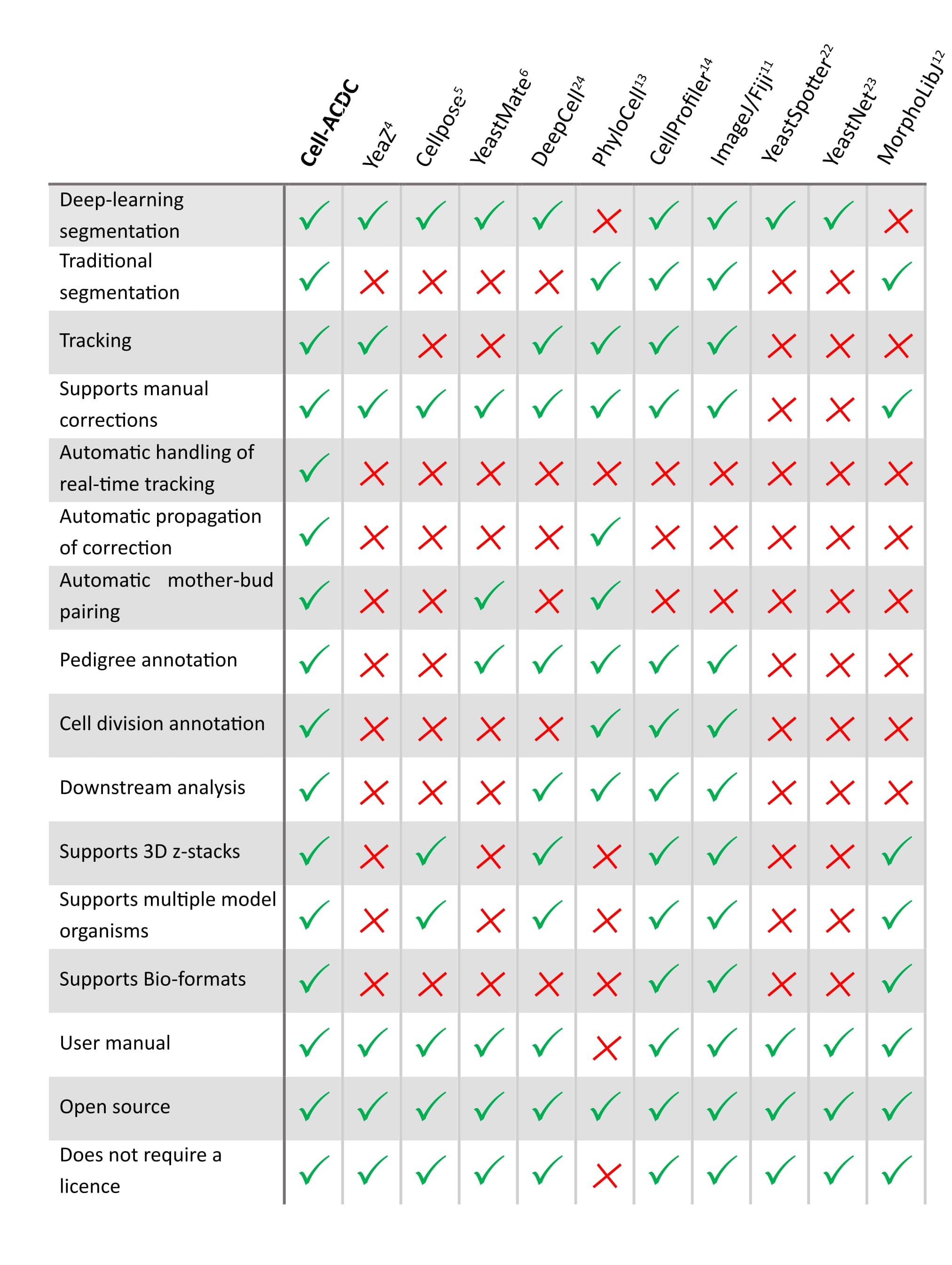
See below how it compares to other popular tools available (Table 1 of our publication).
Of course not! Cell-ACDC automatically computes several single-cell numerical features such as cell area and cell volume, plus the mean, max, median, sum and quantiles of any additional fluorescent channel's signal. It even performs background correction, to compute the protein amount and concentration.
You can load and analyse single 2D images, 3D data (3D z-stacks or 2D images over time) and even 4D data (3D z-stacks over time).
Finally, we provide Jupyter notebooks to visualize and interactively explore the data produced.
Is every second line in your files from your bidirectional microscopy shifted? Look here for further information on how to correct your data.
See here for a list of the scientific publications where Cell-ACDC was used: Link.
- Please find a complete user guide here
- Installation guide
- User manual
- Publication of Cell-ACDC
- Image.sc Forum to ask any question. Make sure to tag the Topic with the tag
cell-acdc. - Report issues, request a feature or ask questions by opening a new issue here
- X thread
- Scientific publications where Cell-ACDC was used
If you find Cell-ACDC useful, please cite it as follows:
Padovani, F., Mairhörmann, B., Falter-Braun, P., Lengefeld, J. & Schmoller, K. M. Segmentation, tracking and cell cycle analysis of live-cell imaging data with Cell-ACDC. BMC Biology 20, 174 (2022). DOI: 10.1186/s12915-022-01372-6
IMPORTANT: when citing Cell-ACDC make sure to also cite the paper of the segmentation models and trackers you used! See here for a list of models currently available in Cell-ACDC.
Do not hesitate to contact us here on GitHub (by opening an issue) or directly at the email [email protected] for any problem and/or feedback on how to improve the user experience!
At Cell-ACDC we encourage contributions to the code! Please read our contributing guide to get started.