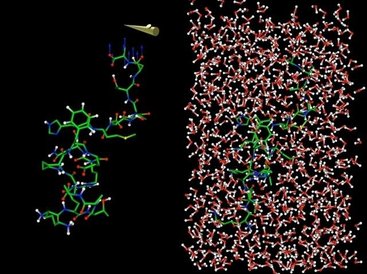
MDDriver is a library allowing a user to easily couple molecular visualisation tools with simulation code through a network. It allows to carry out interactive molecular dynamics, visualize simulation results and drive a simulation in interactive time.
The previous sourceforge repository is now deprecated and the latest version is on github.
This library is needed for:
- Interactive Simulation with Unitymol as a visualisation tool.
- Interactive simulation with the Biospring tool as a simulation engine.
IMD (Interactive Molecular Dynamics) is supported natively in GROMACS and VMD. MDDriver is no longer needed for this kind of simulation. MDDriver is compatible with this IMD protocol used by VMD and GROMACS.
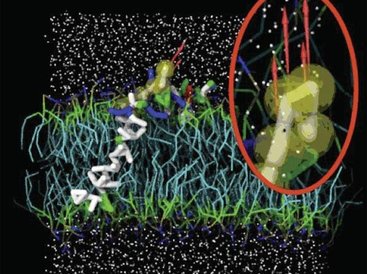
These 3 components (UnityMol, MDDriver and Biospring) have been used together recently to position rapidely and interactively proteins within membranes [1].
- CMake http://www.cmake.org
git clone https://github.com/LBT-CNRS/MDDriver
cd MDDriver/
# On Windows with Microsoft Visual Studio 16 for example
cmake -S . -B build -DCMAKE_INSTALL_PREFIX:PATH=/path/to/mddriver/installation -G "Visual Studio 16 2019"
# On Mac/Linux
cmake -S . -B build -DCMAKE_INSTALL_PREFIX:PATH=/path/to/mddriver/installation
# Compilation
cmake --build build --config Release
cmake --install
By default, the UnityMol plugin is compiled. This can be deactivated with -DBUILD_PLUGINS=OFF.
By default, the example tool in C++ and the python library are not installed. It can be activated with -DINSTALL_EXAMPLE_TOOL=ON and -DINSTALL_PYTHON_LIB=ON.
You can test the protocol by lauching 2 executables, one in each console:
First console:
/path/to/mddriver/installation/bin/servertest
Second console:
/path/to/mddriver/installation/bin/clienttest
You should see energy and coords exchanging by the server and the client.
Usually, MDDriver is used as a library for another code.
For Biospring: give the path to the MDDriver installation when compiling it
For UnityMol: Copy/paste the library file into the Plugins folder of the UnityMol build.
During CI, the UnityMol Plugin is build for all platforms automatically and stored in the artifacts. You can find the latest artifacts in the GitHub Actions page.
[1]: Lanrezac, A.; Laurent, B.; Santuz, H.; Férey, N.; Baaden, M. Fast and Interactive Positioning of Proteins within Membranes. Algorithms 2022, 15, 415. https://doi.org/10.3390/a15110415
MDDriver is licenced under the CeCILL-C license.
- Marc Baaden
- Xavier Martinez
- Hubert Santuz
- André Lanrezac
- Nicolas Ferey
- Olivier Delalande
- Gilles Grasseau