-
Notifications
You must be signed in to change notification settings - Fork 4
Home
In addition to the text based documentation here, additional information in other formats (video, slides) is available at https://umgear.org/manual.html.
The gene Expression Analysis Resource (gEAR) portal is a website for visualization and analysis of multiomic data both in public and private domains. The gEAR enables upload, visualization and analysis of bulk RNA sequencing, scRNA-seq data, and epigenetic data.
gEAR is under active development and we are continuously adding new features. If you need assistance with any issues or have suggestions, please contact our user helpdesk https://umgear.org/contact.html.
The gEAR is maintained by a team at the University of Maryland School of Medicine, and led by Dr. Ronna Hertzano and Joshua Orvis. The gEAR is supported by the NIDCD/NIH R01DC013817, R01DC019370, NIMH/NIH R24MH114815 and the Hearing Health Foundation (Hearing Restoration Project).
The following sections goes over some basic functionality of the gEAR platform.
To create an account, click the create account button at the top right of the homepage or follow the link here (Create gEAR Account). On this page you will enter some basic information about you and your email address (which can be used to recover your password). After creating an account, you will have access to all the tools available on the gEAR. To see your user information, click on your username in the top right.
The gEAR homepage (umgear.org) contains links to the most commonly used tools and utilities on the gEAR. At any time on the site, you can return to the homepage by clicking the gEAR icon in the top left corner.
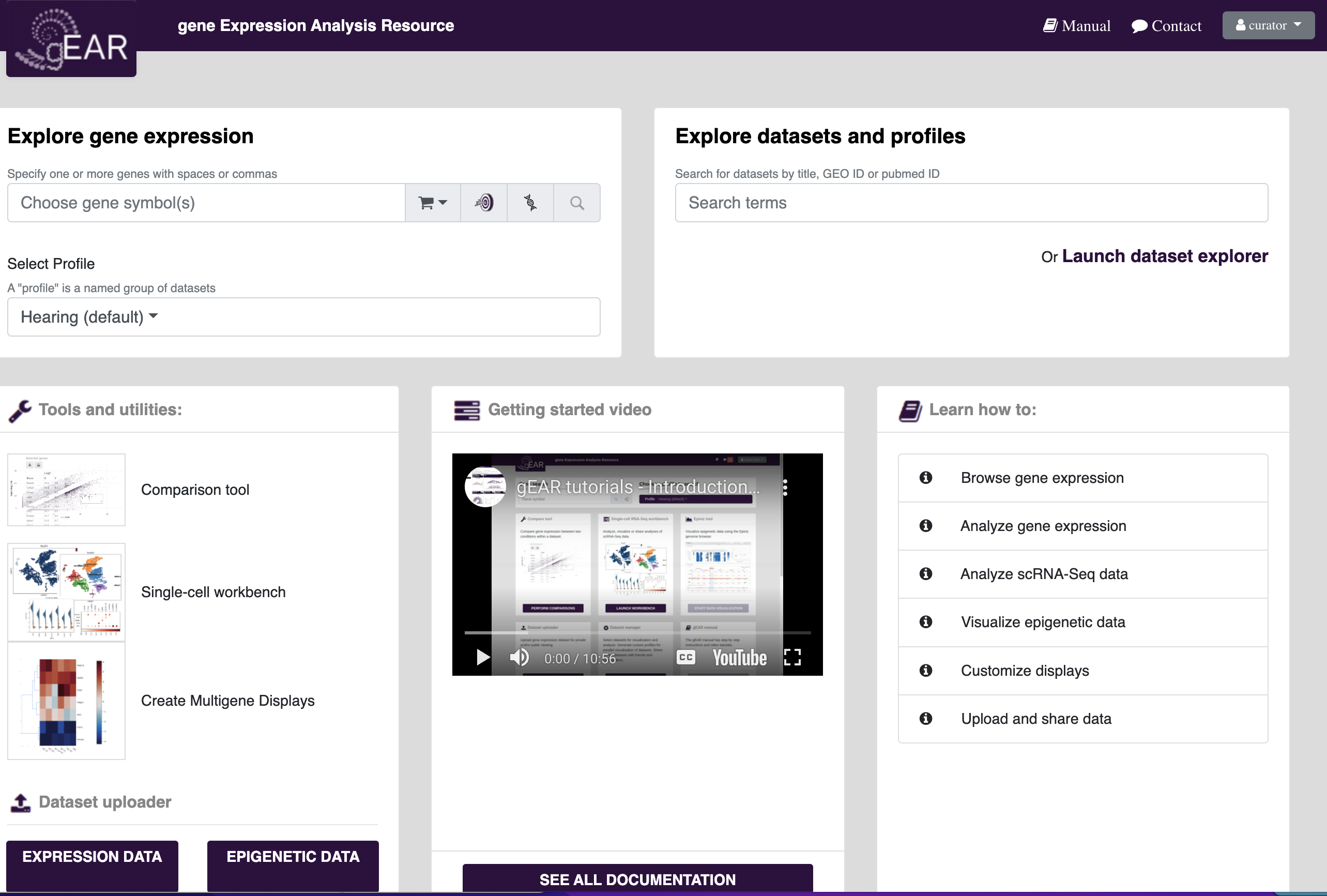
Searching for genes of interest in public datasets is one of the most common uses of the gEAR platform. A good place to start searching is on the homepage. To search for your gene of interest, put your gene name in the gene expression search bar (see below) and press search.
For more information about searching for genes of interest and other options see:
- Walkthrough Slides/Video
- Detailed Walkthrough LINK UPDATED SOON
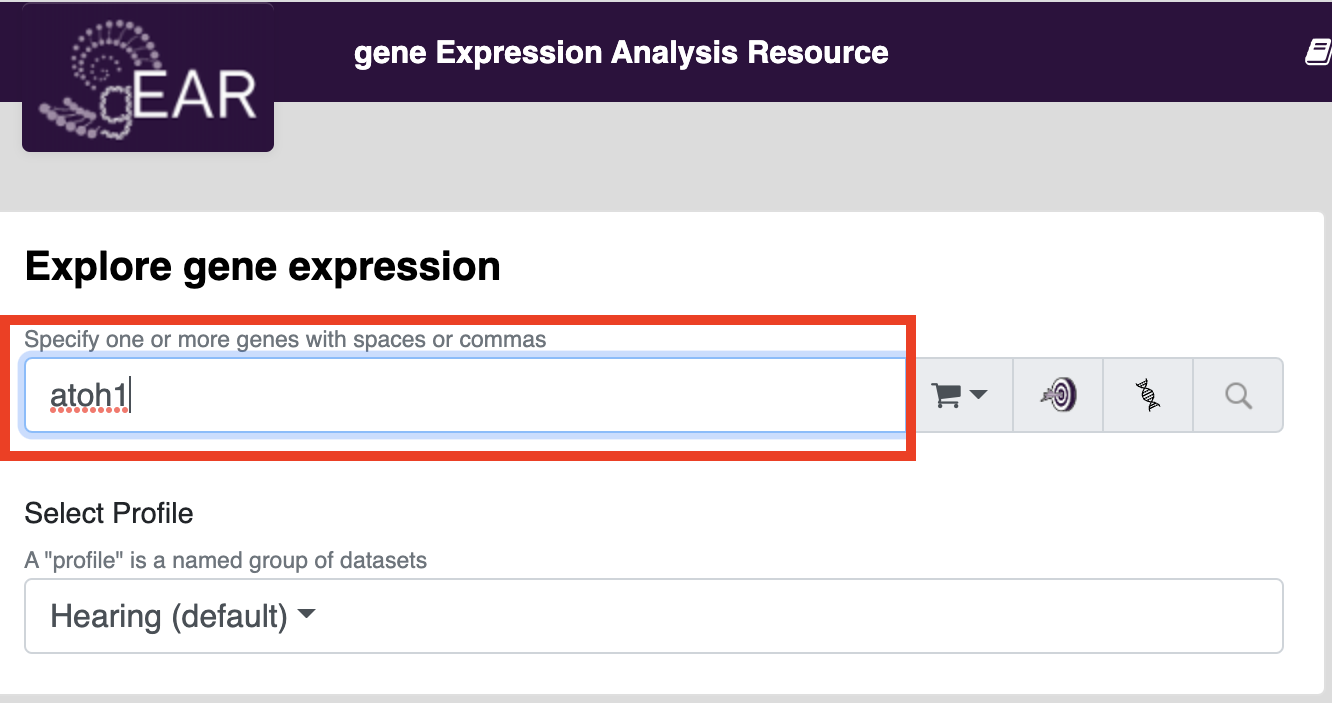
If you wish to look at results for a particular dataset or explore what datasets are available, you can either search by adding terms to the explore datasets box on the homepage or launch the dataset explorer (Either option will open the dataset explorer)

Once in the dataset explorer you can refine your search by type of data, date, organism, etc. by choosing options on the left side of the page. To search for genes within a particular dataset, click on the eye symbol and enter your gene/genes of interest in the search box.
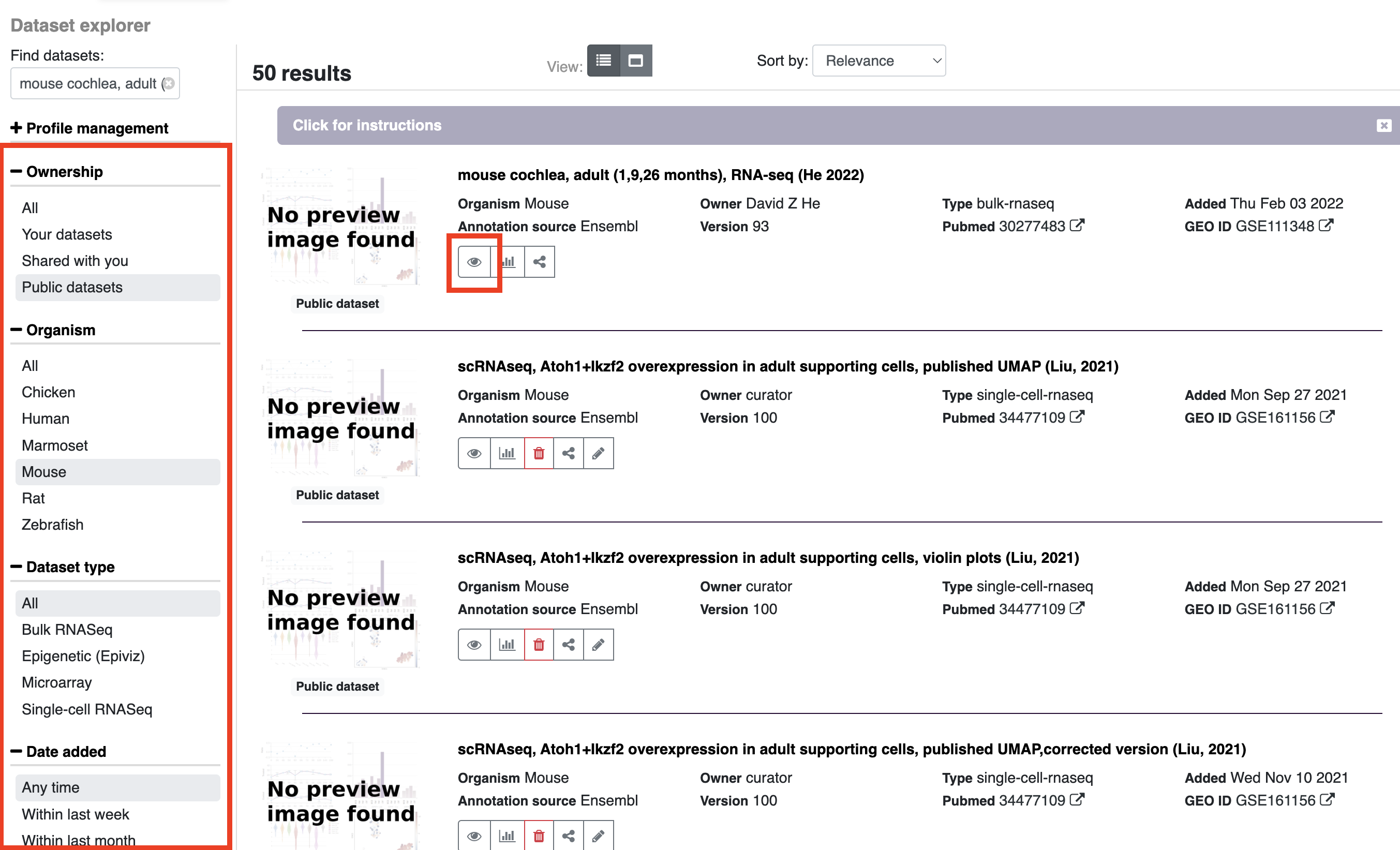
Gene annotations can be viewed on the gEAR platform in several ways. After searching for a gene, you will see several annotation results at the top of the screen.
-
Annotation by organism:
- The icons under this header will provide information about genes across different species. In the example below (searching for Atoh1) currently shows the mouse annotation as highlighted. Choosing other organism icons will provide the name of the gene in that organism (e.g. ATOH1 in humans) and update the external links to match the gene name listed after "Showing results for gene:".
-
External resource links:
- These options open a new window with the pages for the searched gene on other web resources. Note: If your search includes more than one gene, the links will be displayed only for the first gene in your search.
-
Deafness gene annotation:
- On gEAR there is additional annotation information related to deafness (for common deafness related genes). Clicking on the icon will provide a list of phenotypes and links out to deafness specific resources.
-
Functional annotation:
- Clicking on the functional annotation text will expand the window to show additional information for the gene of choice including: GO terms, full names, and any aliases.
Profiles are gEAR's way of collecting together datasets around a similar topic. The gEAR team maintains a number of curated profiles but any user can create their own profile of datasets to explore. For example, the default profile (Hearing) contains 10 datasets that offer a broad overview of gene expression while other profiles may be devoted to specific organisms (e.g. Zebrafish) or collections of displays used in individual publications. Profiles can be created in the Dataset Explorer (CREATING PROFILES UPDATED SOON).
For more information on how to upload different types of data into gEAR, see our upload documentation. Currently gEAR supports the upload of:
- Bulk RNAseq data
- Single Cell sequencing data
- Epigenetic data (Through integration with EpiViz)