2020-03-12 17:04:11
R is a free software environment for statistical computing and graphics. Many users think of R as a statistics system. We prefer to think of it as an environment within which statistical techniques are implemented. However, R is useful for so much more, including advanced data management, programming, and creation of advanced visualizations, both interactive and non-interactive. It’s basically supercharged Excel.
This repository acts as a guide to how ACDI/VOCA staff use R. If you find it useful, please feel free to use it in your own organization.
If you are not an R user and have no idea what any of this means, welcome to the community! (And you’re welcome!)
- Actually, ModernDive wrote an outstanding post on the first steps. Go here and do everything they say!.
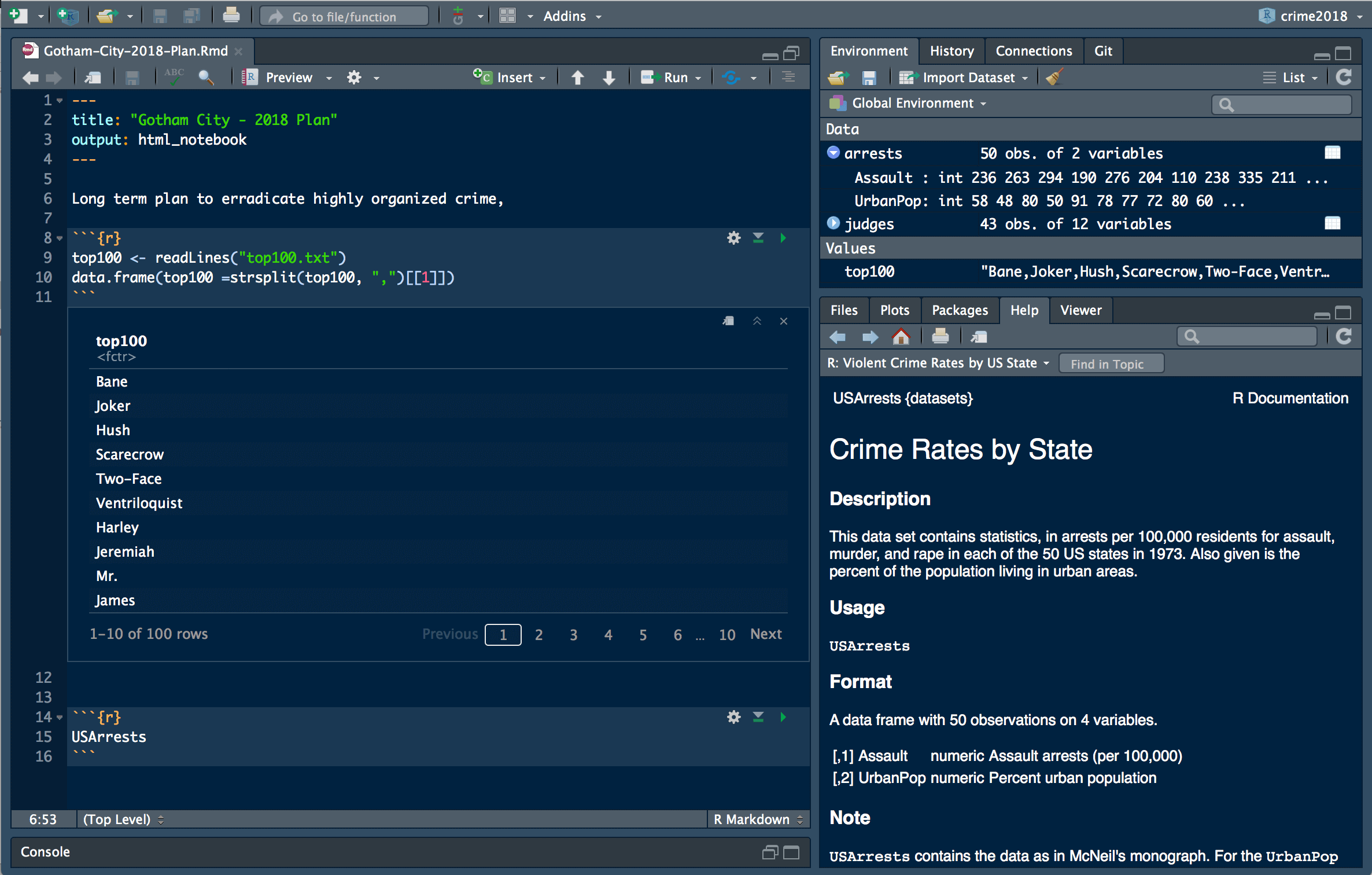
RStudio is an Integrated Development Environment (IDE) for the R programming language and is the IDE of choice for ACDI/VOCA for using R. Split into two editions, RStudio for Desktop and RStudio Server, it is an incredibly empowering tool that allows one to use R to its full potential. The different panels in RStudio such as “Environment”, “Files”, “Help”, “History” allows for easy organization and interacting with all the files and objects in your analysis.
Above is what a RStudio set up might look like. You can spend time to position and size each of the panels however you want along with a great many other aesthetical aspects like the code font and color.
Using R Projects is a vital part of keeping your work separately contained with its own working directory, workspace, history, and source documents. Instead of cluttering up your environment with multiple objects from multiple projects you can have a number of different projects opened in its own window so things from one project won’t interfere with another.
A great resource to get started with R Projects is here.
For a general overview of a project-oriented workflow in R, consult this website (still work-in-progress) here written by Jenny Bryan and Jim Hester from R Studio. For a shorter blogpost version of the above see here.
Now that you have everything organized by using Projects you might want to backup and version control your code and analyses using GitHub. Using GitHub will also allow you to coordinate and collaborate with other analysts on the same project, you will never again have to search your email for “datafile_revised.data” or “final_final_final_draft3.doc”!
A great introduction to switching over to a version control system for your projects can be read here.
For connecting your RStudio Project to Git and GitHub check out this chapter of the book, “Happy Git with R” by Jenny Bryan.
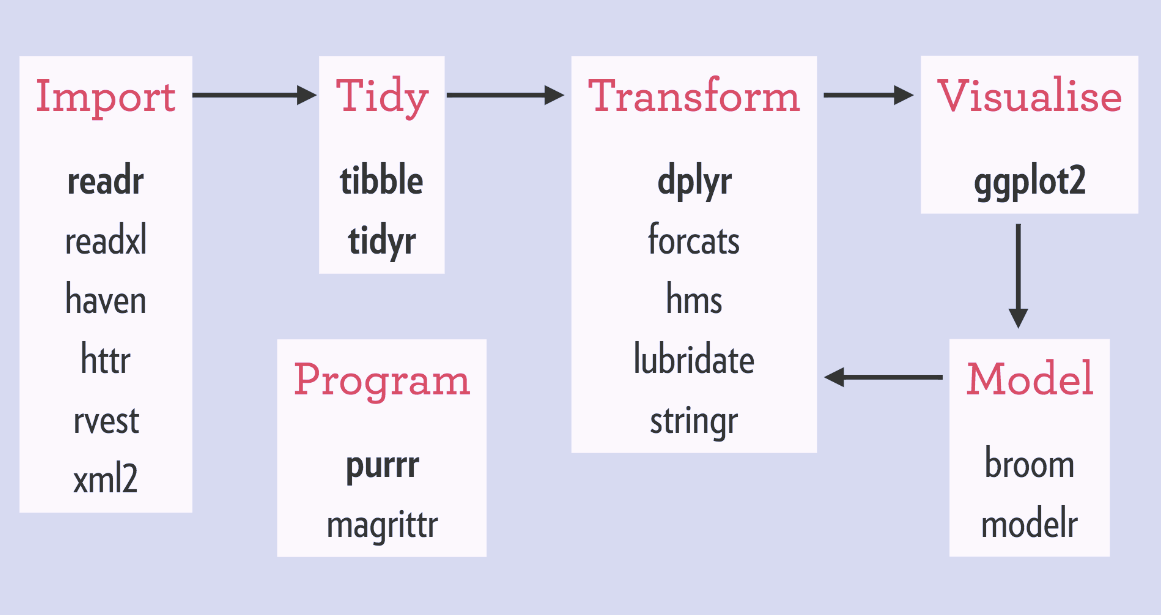
The Tidyverse is a set of R packages specifically designed for doing data science. The packages are consistent in their underlying design and structure which makes it very easy to learn. In regards to syntax and coding style, ACDI VOCA follows the tidyverse style guide for consistent development of our code and tools. One of the main strengths of the tidyverse is that the code is very easily readable by a human compared to base R code. Some great examples of this can be seen here in a great webpage that shows how to do something in both base R and the tidyverse!
The best way to get oriented with the tidyverse packages is to go through Hadley Wickham’s R for Data Science book which is provided online and for free! You can also go visit the Slack group for R4DS here to get online help.
Some particular notes on how we write R code:
- For commenting lines out we use ONE
#whereas if we want to make a comment in the code we use TWO##.
data %>%
select(this, that, everything()) %>%
## Calculate values here:
mutate(nutrition10 = nutrition * 10,
ham_production = ham + production - costs) %>%
## We need to filter by values over 5000:
filter(nutrition10 > 5000) %>%
## No need to arrange by DESC order anymore:
#arrange(desc(nutrition10))- Avoid using
forloops or theapply()family of functions (sapply,lapply,vapply, etc.) and use themap()functions from thepurrrpackage instead. Note that there is nothing wrong with usingforloops in R necessarily but we want to keep everything consistent across the codebase.
library(tidyverse)## Registered S3 method overwritten by 'rvest':
## method from
## read_xml.response xml2
## -- Attaching packages ----------------------------------------------------------------- tidyverse 1.2.1 --
## v ggplot2 3.3.0 v purrr 0.3.3
## v tibble 2.1.3 v dplyr 0.8.4
## v tidyr 1.0.2 v stringr 1.4.0
## v readr 1.3.1 v forcats 0.3.0
## -- Conflicts -------------------------------------------------------------------- tidyverse_conflicts() --
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()
## calculate the mean for 10 observations of a randomly generated normal distribution
1:10 %>%
map(rnorm, n = 10) %>%
map_dbl(mean)## [1] 1.007902 2.097946 3.650947 4.011458 4.652177 5.868114 7.188676
## [8] 8.077154 8.973139 10.166403
- For naming functions and arguments we use snakecase. Ex.
ConnectToMEF()andfieldToMod = "Indicators".
AddThenSubtract <- function(entryNumber, subtractionNumber) {
sum <- entryNumber + 25 - subtractionNumber
sum
}
AddThenSubtract(entryNumber = 5, subtractionNumber = 3.5)## [1] 26.5
There is a tab in R Studio that helps you manage installing packages.
From here you can choose from where you want to install a package,
either from CRAN or a local .tar.gz or .zip file. Then you just type
the name of the package in, be very careful with the proper
capitalization and underscore/dash in the names! Then, make sure you
are installing the package into the proper directory, check whether you
want to install dependencies or not and click install. Remember that
after installing you need to load the package with a library() call
every new session.
If you want to install from the console or in a script:
## Take off thehashtag in the line below to "uncomment" it:
# install.packages("nycflights13")
library(nycflights13)
## let's look at one of the datasets from our newly installed package!
glimpse(airlines)## Observations: 16
## Variables: 2
## $ carrier <chr> "9E", "AA", "AS", "B6", "DL", "EV", "F9", "FL", "HA", ...
## $ name <chr> "Endeavor Air Inc.", "American Airlines Inc.", "Alaska...
Not all packages are available on CRAN, here’s how you would install a package from GitHub:
remotes::install_github("ACDIVOCATech/bulletchartr")
library(bulletchartr)R is an open-source language with tons of community support online and
in print. A great way to read up on the latest R news is to go on
Twitter and search for #rstats. You should also subscribe to R
Bloggers to get a curated stream of news,
blog posts, and tutorials provided by thousands of R users from across
the world.
For getting help, go over to StackOverflow
and look up questions tagged with [r] and any other R package you
might be having trouble with. Another good resource that is solely
dedicated to R problems and discussion is RStudio
Community. One can also try asking for
help on Twitter via the #rstats hashtag as well!
At ACDI/VOCA we have a whole suite of packages that helps us gather and process data from our databases and output them into a variety of dashboards and visualizations. The R Packages book by Hadley Wickham is a great and free online resource for those who want to create their own package to help with their analysis or contribute to one of our own existing packages.
When collaborating on packages (this can apply to projects too) it is important to create your own branch alongside the main stream of development. This allows you and others to work in parallel without overwriting eachother’s work. A good way to organize things is that you leave the main “master” branch alone while everybody
When using an ACDI/VOCA R package, please remember to PULL the latest version from Github from the Github panel in RStudio. When contributing to any of our R packages please create your own branch and when you’re done submit a Pull Request to the package authors. For an introduction on contributing to open-source using GitHub, see here.
Here are some general best practices that should be followed:
- Inputs into complex functions should be stated explicitly, example:
DO THIS:
function(arg1 = x, arg2 = y)instead of this:function(x,y). It’s a bit more annoying, but does help resolve conflicts, especially when working with packages likeACDIVOCAdataGetterthat are continuously in active development.
- Git fetch
- Switch branch to one to test
- Build Package using “BUILD > MORE > CLEAN AND REBUILD”. Alternatively, use R CMD INSTALL –preclean in console.
- Run tests (Ctrl + Shift + T). If this passes, continue
- See if Issue has been resolved. If yes, Merge with Master, and delete branch.
- Pull Master into the server, turn off PT/GQ runners on crontab, run Day_Deleter() and stay vigilant for 30 mins. All should be good.
- Delete local branch (run
git gcorgit pruneorgit branch -D branch_name).
-
Each concept get their own folder starting with "AV_". The functions they depend on are added to a custom package starting with "AVDG".
-
Each concept repository will be kept clean and the root will only contain the runner
R,shandlogfiles. In some cases, auth tokens and other such randomities can be allowed to live on the folder. Everything else should go into these folders (where required):Inputs- Inputs to the process. - BE CAREFUL - sometimes this isn't a real folder, but a symlink from dropbox.Outputs- Whatever comes out of the project. This is almost always symlinked into dropbox.Punctual- one-off scripts, something like testing, or one-time cleans, etc... they all go into this folder
-
Git commit strategy -
Rfiles and theirshrunners should get committed.logfiles, input and output data should not. Counter files (like which inputs have been processed) are in the grey zone... for now let's say yes but I don't like it.